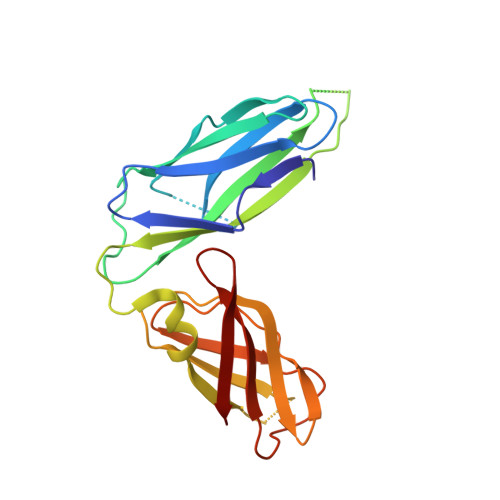
Crystal structure of the usher chaperone YadV reveals a monomer with the proline lock in closed conformation suggestive of an intermediate state.
Pandey, N.K., Verma, G., Kushwaha, G.S., Suar, M., Bhavesh, N.S.(2020) FEBS Lett
- PubMed: 32649775
- DOI: https://doi.org/10.1002/1873-3468.13883
- Primary Citation of Related Structures:
5GHU - PubMed Abstract:
Cell surface pili assembled by the chaperone-usher (CU) pathway play a crucial role in the adhesion of uropathogenic Escherichia coli. YadV is the chaperone component of the CU pathway of Yad pili. Here, we report the crystal structure of YadV from E. coli. In contrast to major usher chaperones, YadV is a monomer in solution as well as in the crystallographic symmetry, and the monomeric form is a preferred state for interacting with pilus subunits. Moreover, we observed a closed conformation for the proline lock, a crucial structural element for chaperone-pilus subunit interaction. MD simulation shows that the closed state of the proline lock is not energetically stable. Thus, the structure of monomeric YadV with its closed proline lock may serve as an intermediate state to provide suitable access to pilus subunits.
- Transcription Regulation Group, International Centre for Genetic Engineering and Biotechnology (ICGEB), New Delhi, India.
Organizational Affiliation: