Structural basis of checkpoint blockade by monoclonal antibodies in cancer immunotherapy
Lee, J.Y., Lee, H.T., Shin, W., Chae, J., Choi, J., Kim, S.H., Lim, H., Won Heo, T., Park, K.Y., Lee, Y.J., Ryu, S.E., Son, J.Y., Lee, J.U., Heo, Y.S.(2016) Nat Commun 7: 13354-13354
- PubMed: 27796306
- DOI: https://doi.org/10.1038/ncomms13354
- Primary Citation of Related Structures:
5GGQ, 5GGR, 5GGS, 5GGT, 5GGU, 5GGV - PubMed Abstract:
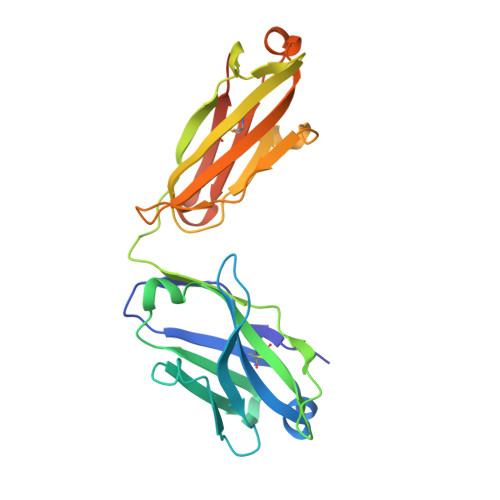
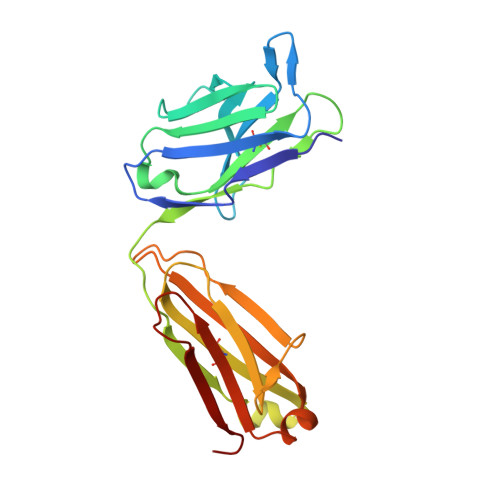
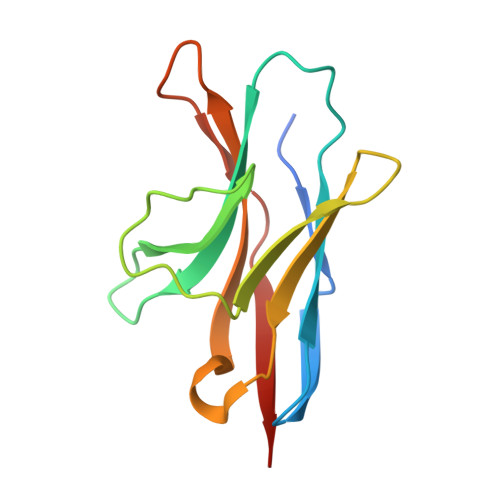
Cancer cells express tumour-specific antigens derived via genetic and epigenetic alterations, which may be targeted by T-cell-mediated immune responses. However, cancer cells can avoid immune surveillance by suppressing immunity through activation of specific inhibitory signalling pathways, referred to as immune checkpoints. In recent years, the blockade of checkpoint molecules such as PD-1, PD-L1 and CTLA-4, with monoclonal antibodies has enabled the development of breakthrough therapies in oncology, and four therapeutic antibodies targeting these checkpoint molecules have been approved by the FDA for the treatment of several types of cancer. Here, we report the crystal structures of checkpoint molecules in complex with the Fab fragments of therapeutic antibodies, including PD-1/pembrolizumab, PD-1/nivolumab, PD-L1/BMS-936559 and CTLA-4/tremelimumab. These complex structures elucidate the precise epitopes of the antibodies and the molecular mechanisms underlying checkpoint blockade, providing useful information for the improvement of monoclonal antibodies capable of attenuating checkpoint signalling for the treatment of cancer.
- Department of Chemistry, Konkuk University, 120 Neungdong-ro, Gwangjin-gu, Seoul 05029, Republic of Korea.
Organizational Affiliation: