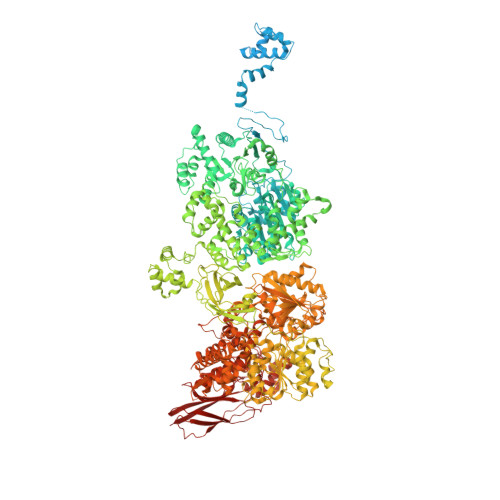
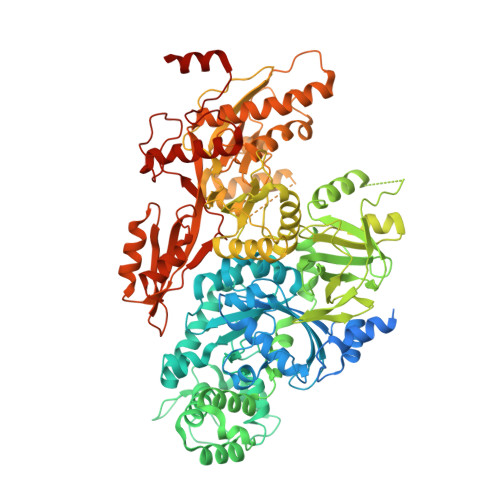
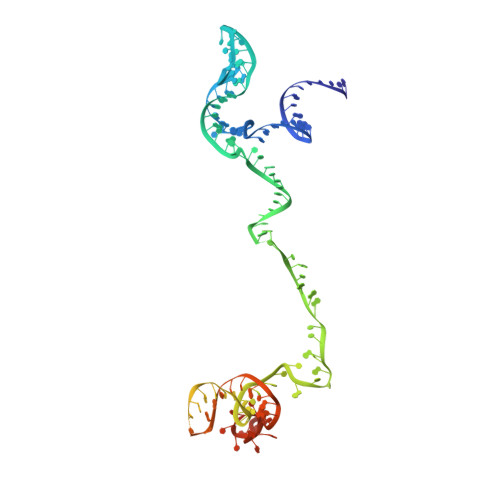
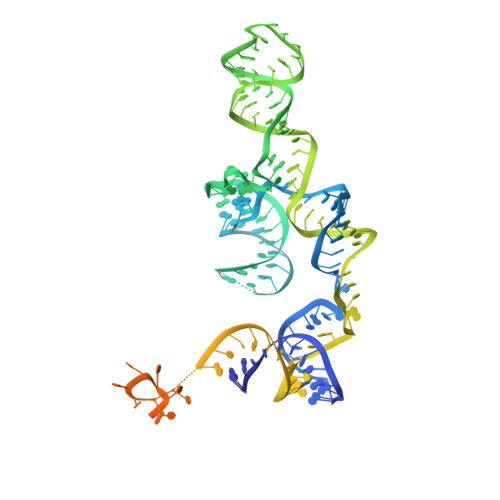
Cryo-EM structure of the yeast U4/U6.U5 tri-snRNP at 3.7 angstrom resolution.
Nguyen, T.H., Galej, W.P., Bai, X.C., Oubridge, C., Newman, A.J., Scheres, S.H., Nagai, K.(2016) Nature 530: 298-302
- PubMed: 26829225
- DOI: https://doi.org/10.1038/nature16940
- Primary Citation of Related Structures:
5GAM, 5GAN, 5GAO, 5GAP - PubMed Abstract:
U4/U6.U5 tri-snRNP represents a substantial part of the spliceosome before activation. A cryo-electron microscopy structure of Saccharomyces cerevisiae U4/U6.U5 tri-snRNP at 3.7 Å resolution led to an essentially complete atomic model comprising 30 proteins plus U4/U6 and U5 small nuclear RNAs (snRNAs). The structure reveals striking interweaving interactions of the protein and RNA components, including extended polypeptides penetrating into subunit interfaces. The invariant ACAGAGA sequence of U6 snRNA, which base-pairs with the 5'-splice site during catalytic activation, forms a hairpin stabilized by Dib1 and Prp8 while the adjacent nucleotides interact with the exon binding loop 1 of U5 snRNA. Snu114 harbours GTP, but its putative catalytic histidine is held away from the γ-phosphate by hydrogen bonding to a tyrosine in the amino-terminal domain of Prp8. Mutation of this histidine to alanine has no detectable effect on yeast growth. The structure provides important new insights into the spliceosome activation process leading to the formation of the catalytic centre.
- MRC Laboratory of Molecular Biology Francis Crick Avenue Cambridge CB2 0QH UK.
Organizational Affiliation: