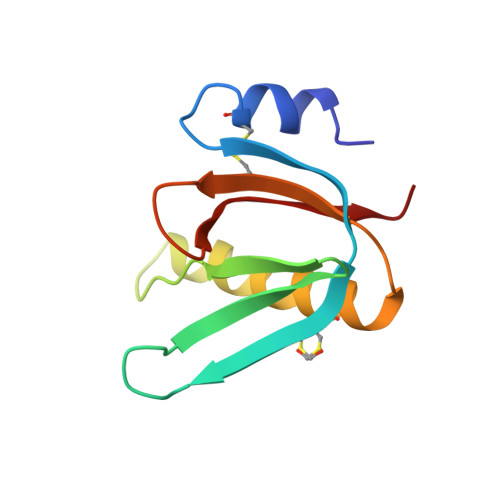
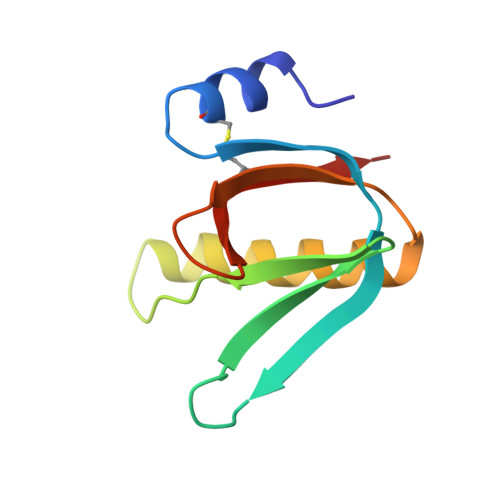
Biochemical and Structural Characterization of the Novel Sialic Acid-Binding Site of Escherichia Coli Heat-Labile Enterotoxin Lt-Iib.
Zalem, D., Ribeiro, J.P., Varrot, A., Lebens, M., Imberty, A., Teneberg, S.(2016) Biochem J 473: 3923
- PubMed: 27562297
- DOI: https://doi.org/10.1042/BCJ20160575
- Primary Citation of Related Structures:
5G3L - PubMed Abstract:
The structurally related AB 5 -type heat-labile enterotoxins of Escherichia coli and Vibrio cholerae are classified into two major types. The type I group includes cholera toxin (CT) and E. coli LT-I, whereas the type II subfamily comprises LT-IIa, LT-IIb and LT-IIc. The carbohydrate-binding specificities of LT-IIa, LT-IIb and LT-IIc are distinctive from those of cholera toxin and E. coli LT-I. Whereas CT and LT-I bind primarily to the GM1 ganglioside, LT-IIa binds to gangliosides GD1a, GD1b and GM1, LT-IIb binds to the GD1a and GT1b gangliosides, and LT-IIc binds to GM1, GM2, GM3 and GD1a. These previous studies of the binding properties of type II B-subunits have been focused on ganglio core chain gangliosides. To further define the carbohydrate binding specificity of LT-IIb B-subunits, we have investigated its binding to a collection of gangliosides and non-acid glycosphingolipids with different core chains. A high-affinity binding of LT-IIb B-subunits to gangliosides with a neolacto core chain, such as Neu5Gcα3- and Neu5Acα3-neolactohexaosylceramide, and Neu5Gcα3- and Neu5Acα3-neolactooctaosylceramide was detected. An LT-IIb-binding ganglioside was isolated from human small intestine and characterized as Neu5Acα3-neolactohexaosylceramide. The crystal structure of the B-subunit of LT-IIb with the pentasaccharide moiety of Neu5Acα3-neolactotetraosylceramide (Neu5Ac-nLT: Neu5Acα3Galβ4GlcNAcβ3Galβ4Glc) was determined providing the first information for a sialic-binding site in this subfamily, with clear differences from that of CT and LT-I.
- Institute of Biomedicine, Department of Medical Biochemistry and Cell Biology, The Sahlgrenska Academy, University of Gothenburg, Göteborg, Sweden.
Organizational Affiliation: