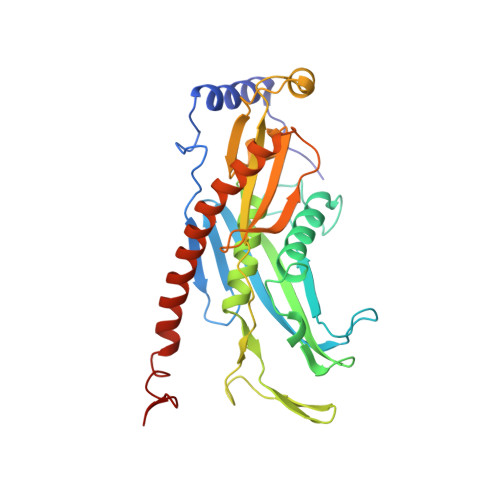
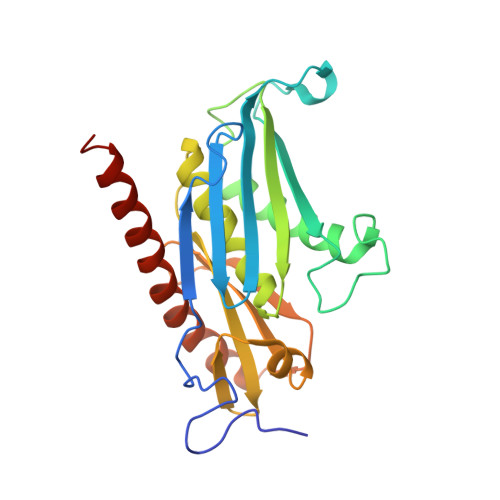
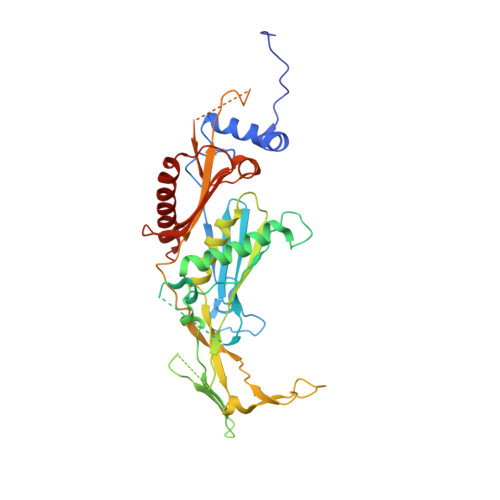
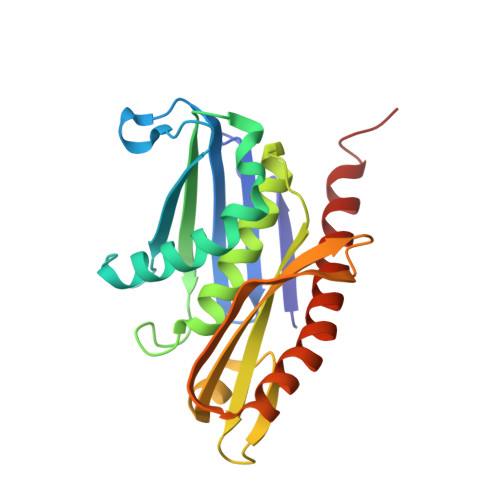
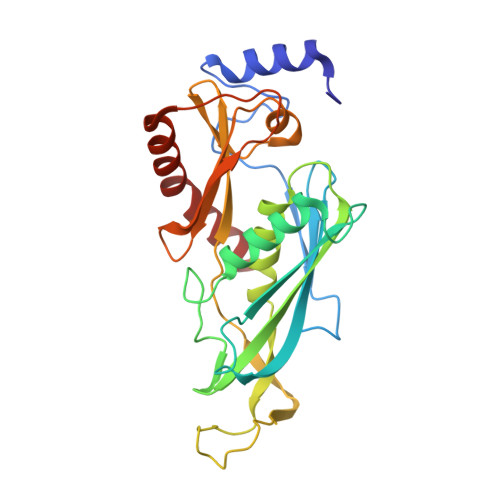
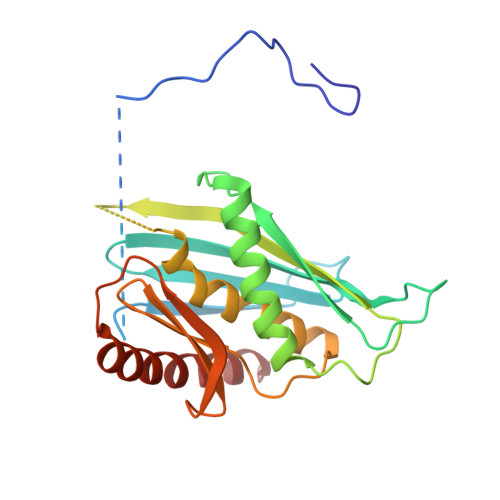
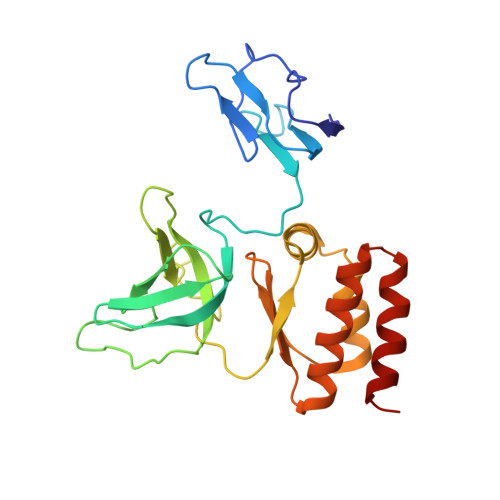
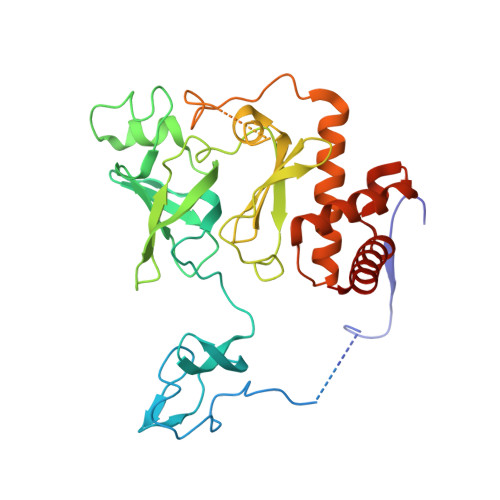
Cryoem Structure of Yeast Cytoplasmic Exosome Complex.
Liu, J., Niu, C., Wu, Y., Tan, D., Wang, Y., Ye, M., Liu, Y., Zhao, W., Zhou, K., Liu, Q., Dai, J., Yang, X., Dong, M., Huang, N., Wang, H.(2016) Cell Res 26: 822
- PubMed: 27174052
- DOI: https://doi.org/10.1038/cr.2016.56
- Primary Citation of Related Structures:
5G06 - PubMed Abstract:
The eukaryotic multi-subunit RNA exosome complex plays crucial roles in 3'-to-5' RNA processing and decay. Rrp6 and Ski7 are the major cofactors for the nuclear and cytoplasmic exosomes, respectively. In the cytoplasm, Ski7 helps the exosome to target mRNAs for degradation and turnover via a through-core pathway. However, the interaction between Ski7 and the exosome complex has remained unclear. The transaction of RNA substrates within the exosome is also elusive. In this work, we used single-particle cryo-electron microscopy to solve the structures of the Ski7-exosome complex in RNA-free and RNA-bound forms at resolutions of 4.2 Å and 5.8 Å, respectively. These structures reveal that the N-terminal domain of Ski7 adopts a structural arrangement and interacts with the exosome in a similar fashion to the C-terminal domain of nuclear Rrp6. Further structural analysis of exosomes with RNA substrates harboring 3' overhangs of different length suggests a switch mechanism of RNA-induced exosome activation in the through-core pathway of RNA processing.
- Ministry of Education Key Laboratory of Protein Sciences, Tsinghua-Peking Joint Center for Life Sciences, Beijing Advanced Innovation Center for Structural Biology, School of Life Sciences, Tsinghua University, Beijing 100084, China.
Organizational Affiliation: