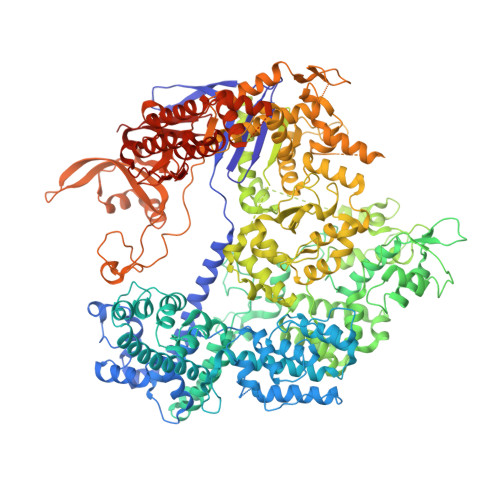
Structural Plasticity of Pam Recognition by Engineered Variants of the RNA-Guided Endonuclease Cas9.
Anders, C., Bargsten, K., Jinek, M.(2016) Mol Cell 61: 895
- PubMed: 26990992
- DOI: https://doi.org/10.1016/j.molcel.2016.02.020
- Primary Citation of Related Structures:
5FW1, 5FW2, 5FW3 - PubMed Abstract:
The RNA-guided endonuclease Cas9 from Streptococcus pyogenes (SpCas9) forms the core of a powerful genome editing technology. DNA cleavage by SpCas9 is dependent on the presence of a 5'-NGG-3' protospacer adjacent motif (PAM) in the target DNA, restricting the choice of targetable sequences. To address this limitation, artificial SpCas9 variants with altered PAM specificities have recently been developed. Here we report crystal structures of the VQR, EQR, and VRER SpCas9 variants bound to target DNAs containing their preferred PAM sequences. The structures reveal that the non-canonical PAMs are recognized by an induced fit mechanism. Besides mediating sequence-specific base recognition, the amino acid substitutions introduced in the SpCas9 variants facilitate conformational remodeling of the PAM region of the bound DNA. Guided by the structural data, we engineered a SpCas9 variant that specifically recognizes NAAG PAMs. Taken together, these studies inform further development of Cas9-based genome editing tools.
- Department of Biochemistry, University of Zurich, Winterthurerstrasse 190, CH-8057 Zurich, Switzerland.
Organizational Affiliation: