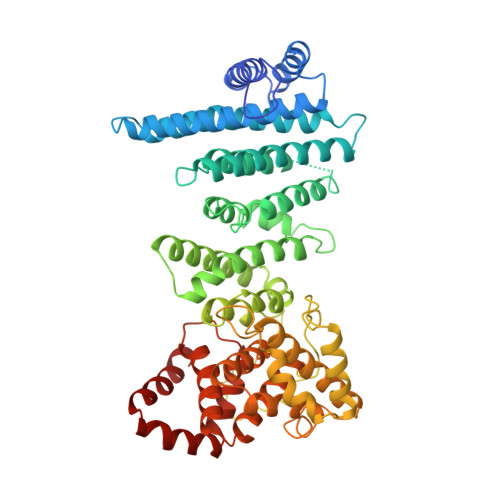
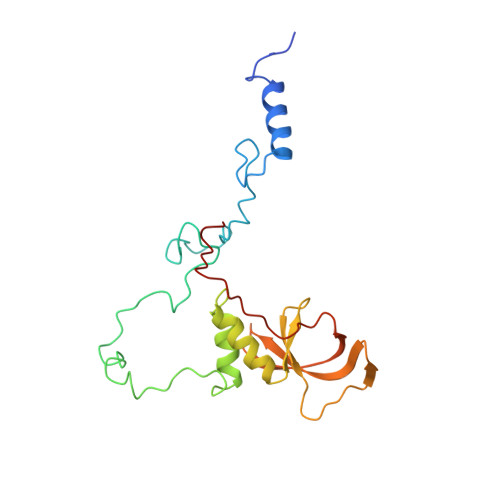
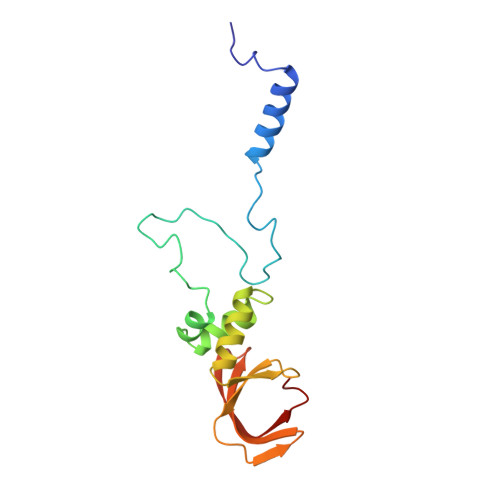
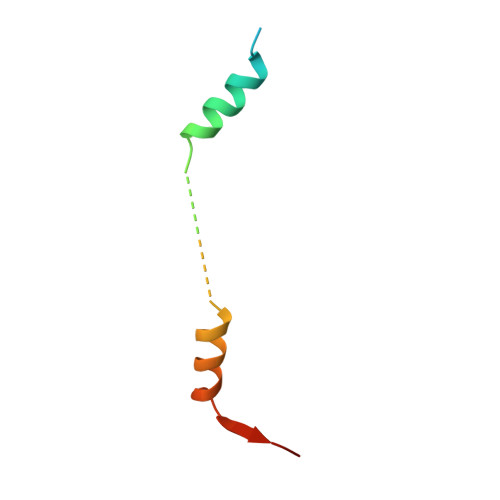
Distinct Modes of Recruitment of the Ccr4-not Complex by Drosophila and Vertebrate Nanos
Raisch, T., Bhandari, D., Sabath, K., Helms, S., Valkov, E., Weichenrieder, O., Izaurralde, E.(2016) EMBO J 35: 974
- PubMed: 26968986
- DOI: https://doi.org/10.15252/embj.201593634
- Primary Citation of Related Structures:
5FU6, 5FU7 - PubMed Abstract:
Nanos proteins repress the expression of target mRNAs by recruiting effector complexes through non-conserved N-terminal regions. In vertebrates, Nanos proteins interact with the NOT1 subunit of the CCR4-NOT effector complex through a NOT1 interacting motif (NIM), which is absent in Nanos orthologs from several invertebrate species. Therefore, it has remained unclear whether the Nanos repressive mechanism is conserved and whether it also involves direct interactions with the CCR4-NOT deadenylase complex in invertebrates. Here, we identify an effector domain (NED) that is necessary for the Drosophila melanogaster (Dm) Nanos to repress mRNA targets. The NED recruits the CCR4-NOT complex through multiple and redundant binding sites, including a central region that interacts with the NOT module, which comprises the C-terminal domains of NOT1-3. The crystal structure of the NED central region bound to the NOT module reveals an unanticipated bipartite binding interface that contacts NOT1 and NOT3 and is distinct from the NIM of vertebrate Nanos. Thus, despite the absence of sequence conservation, the N-terminal regions of Nanos proteins recruit CCR4-NOT to assemble analogous repressive complexes.
- Department of Biochemistry, Max Planck Institute for Developmental Biology, Tübingen, Germany.
Organizational Affiliation: