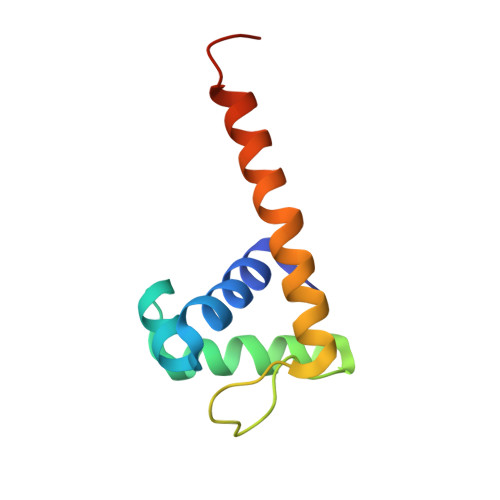
Structural Basis for DNA Recognition by the Transcription Regulator Metr.
Punekar, A.S., Porter, J., Carr, S.B., Phillips, S.E.V.(2016) Acta Crystallogr F Struct Biol Commun 72: 417
- PubMed: 27303893
- DOI: https://doi.org/10.1107/S2053230X16006828
- Primary Citation of Related Structures:
5FO5 - PubMed Abstract:
MetR, a LysR-type transcriptional regulator (LTTR), has been extensively studied owing to its role in the control of methionine biosynthesis in proteobacteria. A MetR homodimer binds to a 24-base-pair operator region of the met genes and specifically recognizes the interrupted palindromic sequence 5'-TGAA-N5-TTCA-3'. Mechanistic details underlying the interaction of MetR with its target DNA at the molecular level remain unknown. In this work, the crystal structure of the DNA-binding domain (DBD) of MetR was determined at 2.16 Å resolution. MetR-DBD adopts a winged-helix-turn-helix (wHTH) motif and shares significant fold similarity with the DBD of the LTTR protein BenM. Furthermore, a data-driven macromolecular-docking strategy was used to model the structure of MetR-DBD bound to DNA, which revealed that a bent conformation of DNA is required for the recognition helix α3 and the wing loop of the wHTH motif to interact with the major and minor grooves, respectively. Comparison of the MetR-DBD-DNA complex with the crystal structures of other LTTR-DBD-DNA complexes revealed residues that may confer operator-sequence binding specificity for MetR. Taken together, the results show that MetR-DBD uses a combination of direct base-specific interactions and indirect shape recognition of the promoter to regulate the transcription of met genes.
- Research Complex at Harwell, Rutherford Appleton Laboratory, Harwell Oxford, Didcot OX11 0FA, England.
Organizational Affiliation: