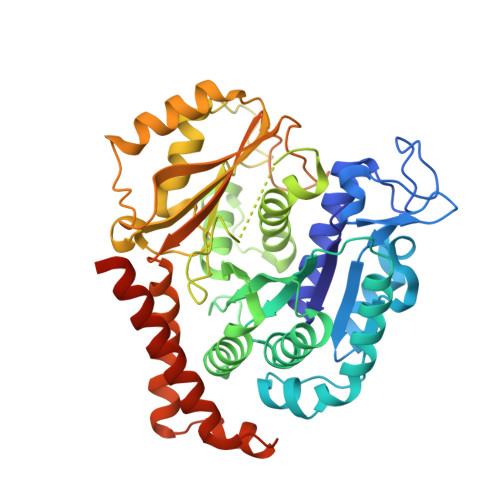
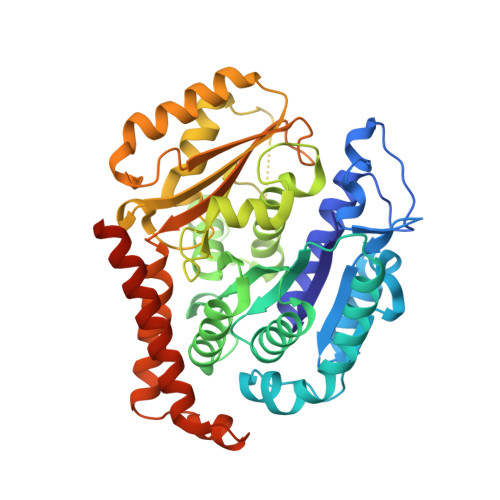
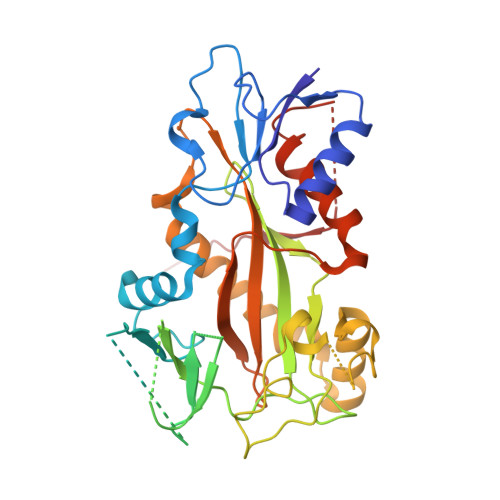
Pironetin Reacts Covalently with Cysteine-316 of Alpha-Tubulin to Destabilize Microtubule.
Yang, J., Wang, Y., Wang, T., Jiang, J., Botting, C.H., Liu, H., Chen, Q., Yang, J., Naismith, J.H., Zhu, X., Chen, L.(2016) Nat Commun 7: 12103
- PubMed: 27357539
- DOI: https://doi.org/10.1038/ncomms12103
- Primary Citation of Related Structures:
5FNV, 5JQG - PubMed Abstract:
Molecules that alter the normal dynamics of microtubule assembly and disassembly include many anticancer drugs in clinical use. So far all such therapeutics target β-tubulin, and structural biology has explained the basis of their action and permitted design of new drugs. However, by shifting the profile of β-tubulin isoforms, cancer cells become resistant to treatment. Compounds that bind to α-tubulin are less well characterized and unexploited. The natural product pironetin is known to bind to α-tubulin and is a potent inhibitor of microtubule polymerization. Previous reports had identified that pironetin reacts with lysine-352 residue however analogues designed on this model had much lower potency, which was difficult to explain, hindering further development. We report crystallographic and mass spectrometric data that reveal that pironetin forms a covalent bond to cysteine-316 in α-tubulin via a Michael addition reaction. These data provide a basis for the rational design of α-tubulin targeting chemotherapeutics.
- State Key Laboratory of Biotherapy and Cancer Center, West China Hospital of Sichuan University and Collaborative Innovation Center of Biotherapy and Cancer, Chengdu 610041, China.
Organizational Affiliation: