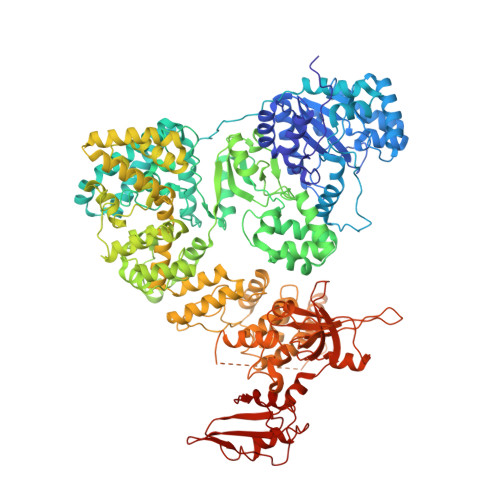
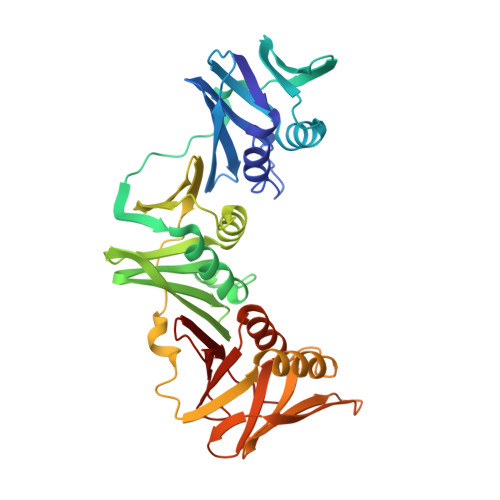
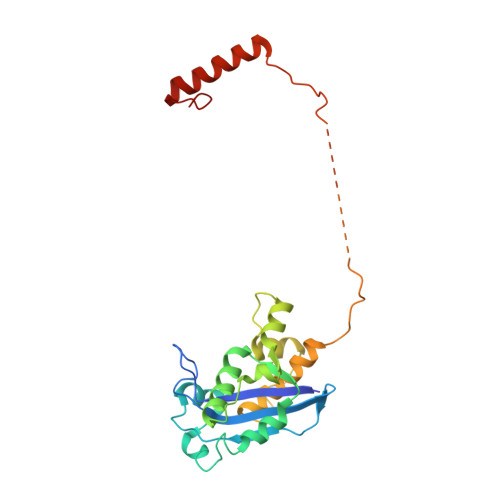
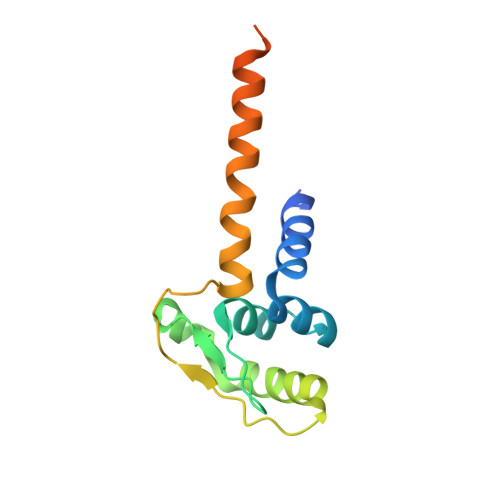
cryo-EM structures of theE. colireplicative DNA polymerase reveal its dynamic interactions with the DNA sliding clamp, exonuclease andtau.
Fernandez-Leiro, R., Conrad, J., Scheres, S.H., Lamers, M.H.(2015) Elife 4
- PubMed: 26499492
- DOI: https://doi.org/10.7554/eLife.11134
- Primary Citation of Related Structures:
5FKU, 5FKV, 5FKW - PubMed Abstract:
The replicative DNA polymerase PolIIIα from Escherichia coli is a uniquely fast and processive enzyme. For its activity it relies on the DNA sliding clamp β, the proofreading exonuclease ε and the C-terminal domain of the clamp loader subunit τ. Due to the dynamic nature of the four-protein complex it has long been refractory to structural characterization. Here we present the 8 Å resolution cryo-electron microscopy structures of DNA-bound and DNA-free states of the PolIII-clamp-exonuclease-τ c complex. The structures show how the polymerase is tethered to the DNA through multiple contacts with the clamp and exonuclease. A novel contact between the polymerase and clamp is made in the DNA bound state, facilitated by a large movement of the polymerase tail domain and τ c . These structures provide crucial insights into the organization of the catalytic core of the replisome and form an important step towards determining the structure of the complete holoenzyme.
- MRC Laboratory of Molecular Biology, Cambridge, United Kingdom.
Organizational Affiliation: