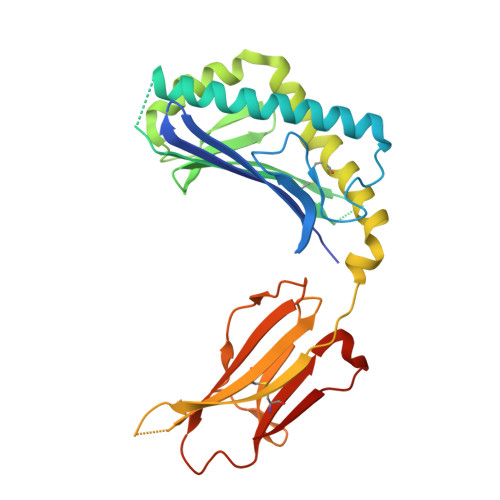
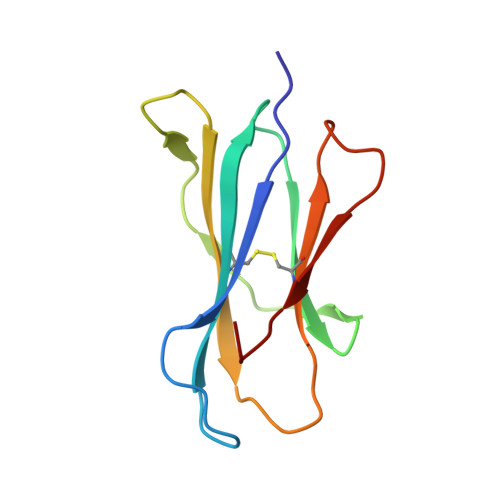
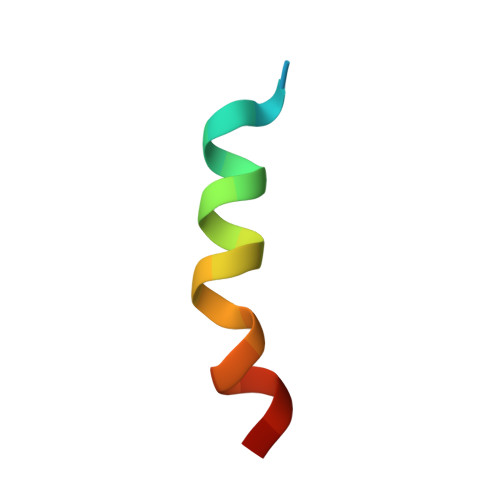
Structure of an Alpha-Helical Peptide and Lipopeptide Bound to the Non-Classical Mhc Class I Molecule Cd1D
Girardi, E., Wang, J., Zajonc, D.M.(2016) J Biological Chem 291: 10677
- PubMed: 27006394
- DOI: https://doi.org/10.1074/jbc.M115.702118
- Primary Citation of Related Structures:
5EFI, 5FKP - PubMed Abstract:
Mouse CD1d is a nonclassical MHC molecule able to present lipids and glycolipids to a specialized subset of T cells known as natural killer T cells. The antigens presented by CD1d have been shown to cover a broad range of chemical structures and to follow precise rules determining the potency of the antigen in the context of T cell activation. Together with lipids, initial reports suggested that CD1d can also bind and present hydrophobic peptides with (F/W)XX(I/L/M)XXW. However, the exact location of peptide binding and the molecular basis for the required motif are currently unknown. Here we present the crystal structure of the first peptide identified to bind CD1d, p99, and show that it binds in the antigen-binding groove of CD1d in a manner compatible with its presentation to T cell receptors. Interestingly, the peptide adopts an α-helical conformation, which orients the motif residues toward its deep binding groove, therefore explaining the molecular requirements for peptide binding. Moreover, we demonstrate that a lipopeptide version of the same peptide is able to bind CD1d in a similar conformation, identifying another class of molecules binding this antigen-presenting molecule.
- From the Division of Cell Biology, La Jolla Institute for Allergy and Immunology, La Jolla, California 92037 and.
Organizational Affiliation: