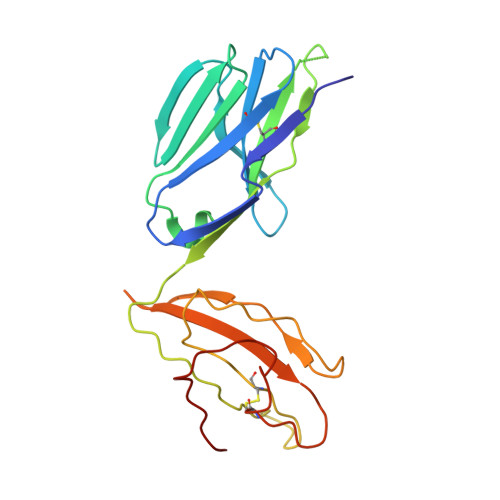
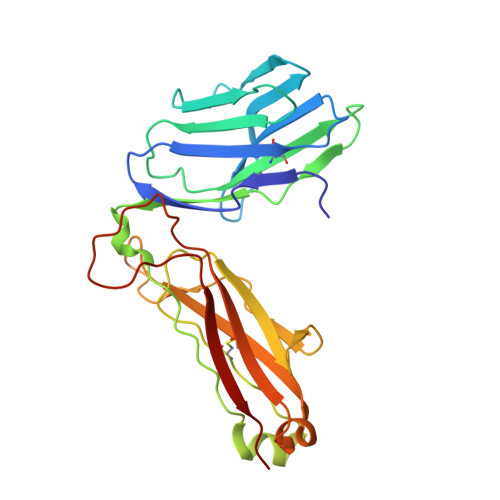
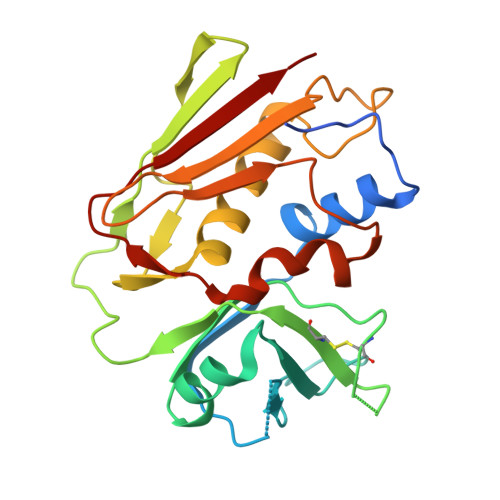
Two Common Structural Motifs for Tcr Recognition by Staphylococcal Enterotoxins
Rodstrom, K.E.J., Regenthal, P., Bahl, C., Ford, A., Baker, D., Lindkvist-Petersson, K.(2016) Sci Rep 6: 25796
- PubMed: 27180909
- DOI: https://doi.org/10.1038/srep25796
- Primary Citation of Related Structures:
5FK9, 5FKA - PubMed Abstract:
Superantigens are toxins produced by Staphylococcus aureus, called staphylococcal enterotoxins (abbreviated SEA to SEU). They can cross-link the T cell receptor (TCR) and major histocompatibility complex class II, triggering a massive T cell activation and hence disease. Due to high stability and toxicity, superantigens are potential agents of bioterrorism. Hence, antagonists may not only be useful in the treatment of disease but also serve as countermeasures to biological warfare. Of particular interest are inhibitors against SEA and SEB. SEA is the main cause of food poisoning, while SEB is a common toxin manufactured as a biological weapon. Here, we present the crystal structures of SEA in complex with TCR and SEE in complex with the same TCR, complemented with computational alanine-scanning mutagenesis of SEA, SEB, SEC3, SEE, and SEH. We have identified two common areas that contribute to the general TCR binding for these superantigens. This paves the way for design of single antagonists directed towards multiple toxins.
- Department of Experimental Medical Science, Lund University, BMC C13, 22 184, Lund, Sweden.
Organizational Affiliation: