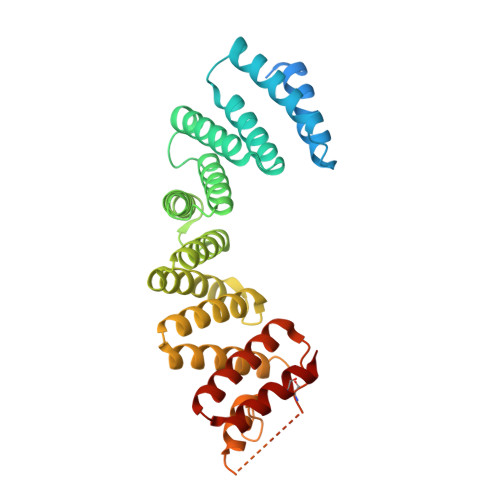The Light Chains of Kinesin-1 are Autoinhibited.
Yip, Y.Y., Pernigo, S., Sanger, A., Xu, M., Parsons, M., Steiner, R.A., Dodding, M.P.(2016) Proc Natl Acad Sci U S A 113: 2418
- PubMed: 26884162
- DOI: https://doi.org/10.1073/pnas.1520817113
- Primary Citation of Related Structures:
5FJY - PubMed Abstract:
The light chains (KLCs) of the microtubule motor kinesin-1 bind cargoes and regulate its activity. Through their tetratricopeptide repeat domain (KLC(TPR)), they can recognize short linear peptide motifs found in many cargo proteins characterized by a central tryptophan flanked by aspartic/glutamic acid residues (W-acidic). Using a fluorescence resonance energy transfer biosensor in combination with X-ray crystallographic, biochemical, and biophysical approaches, we describe how an intramolecular interaction between the KLC2(TPR) domain and a conserved peptide motif within an unstructured region of the molecule, partly occludes the W-acidic binding site on the TPR domain. Cargo binding displaces this interaction, effecting a global conformational change in KLCs resulting in a more extended conformation. Thus, like the motor-bearing kinesin heavy chains, KLCs exist in a dynamic conformational state that is regulated by self-interaction and cargo binding. We propose a model by which, via this molecular switch, W-acidic cargo binding regulates the activity of the holoenzyme.
- Randall Division of Cell and Molecular Biophysics, King's College London, London SE1 1UL, United Kingdom.
Organizational Affiliation: