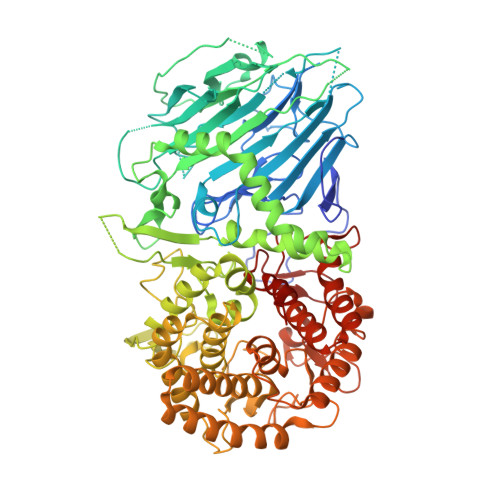
Bacterial Beta-Glucosidase Reveals the Structural and Functional Basis of Genetic Defects in Human Glucocerebrosidase 2 (Gba2)
Charoenwattanasatien, R., Pengthaisong, S., Breen, I., Mutoha, R., Sansenya, S., Hua, Y., Tankrathok, A., Wu, L., Songsiriritthigul, C., Tanaka, H., Williams, S.J., Davies, G.J., Kurisu, G., Ketudat Cairns, J.R.(2016) ACS Chem Biol 11: 1891
- PubMed: 27115290
- DOI: https://doi.org/10.1021/acschembio.6b00192
- Primary Citation of Related Structures:
5BVU, 5BX2, 5BX3, 5BX4, 5BX5, 5FJS - PubMed Abstract:
Human glucosylcerebrosidase 2 (GBA2) of the CAZy family GH116 is responsible for the breakdown of glycosphingolipids on the cytoplasmic face of the endoplasmic reticulum and Golgi apparatus. Genetic defects in GBA2 result in spastic paraplegia and cerebellar ataxia, while cross-talk between GBA2 and GBA1 glucosylceramidases may affect Gaucher disease. Here, we report the first three-dimensional structure for any GH116 enzyme, Thermoanaerobacterium xylanolyticum TxGH116 β-glucosidase, alone and in complex with diverse ligands. These structures allow identification of the glucoside binding and active site residues, which are shown to be conserved with GBA2. Mutagenic analysis of TxGH116 and structural modeling of GBA2 provide a detailed structural and functional rationale for pathogenic missense mutations of GBA2.
- Institute for Protein Research, Osaka University , 3-2 Yamadaoka, Suita, Osaka 565-0871, Japan.
Organizational Affiliation: