Crystal Structure of Raptor Adenovirus 1 Fibre Head and Role of the Beta-Hairpin in Siadenovirus Fibre Head Domains
Nguyen, T.H., Ballmann, M.Z., Do, H.T., Truong, H.N., Benko, M., Harrach, B., van Raaij, M.J.(2016) Virol J 13: 106
- PubMed: 27334597
- DOI: https://doi.org/10.1186/s12985-016-0558-7
- Primary Citation of Related Structures:
5FJL, 5FLD - PubMed Abstract:
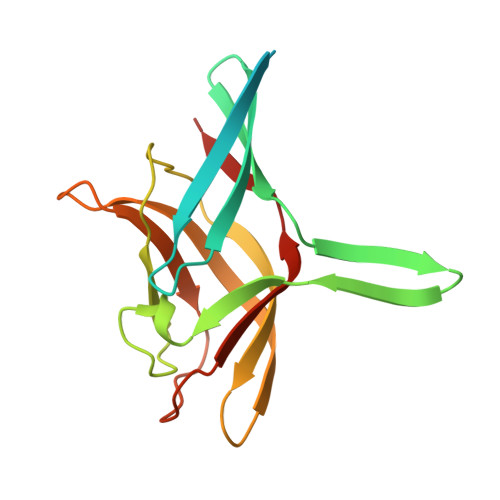
Most adenoviruses recognize their host cells via an interaction of their fibre head domains with a primary receptor. The structural framework of adenovirus fibre heads is conserved between the different adenovirus genera for which crystal structures have been determined (Mastadenovirus, Aviadenovirus, Atadenovirus and Siadenovirus), but genus-specific differences have also been observed. The only known siadenovirus fibre head structure, that of turkey adenovirus 3 (TAdV-3), revealed a twisted beta-sandwich resembling the reovirus fibre head architecture more than that of other adenovirus fibre heads, plus a unique beta-hairpin embracing a neighbouring monomer. The TAdV-3 fibre head was shown to bind sialyllactose. Raptor adenovirus 1 (RAdV-1) fibre head was expressed, crystallized and its structure was solved and refined at 1.5 Å resolution. The structure could be solved by molecular replacement using the TAdV-3 fibre head structure as a search model, despite them sharing a sequence identity of only 19 %. Versions of both the RAdV-1 and TAdV-3 fibre heads with their beta-hairpin arm deleted were prepared and their stabilities were compared with the non-mutated proteins by a thermal unfolding assay. The structure of the RAdV-1 fibre head contains the same twisted ABCJ-GHID beta-sandwich and beta-hairpin arm as the TAdV-3 fibre head. However, while the predicted electro-potential surface charge of the TAdV-3 fibre head is mainly positive, the RAdV-1 fibre head shows positively and negatively charged patches and does not appear to bind sialyllactose. Deletion of the beta-hairpin arm does not affect the structure of the raptor adenovirus 1 fibre head and only affects the stability of the RAdV-1 and TAdV-3 fibre heads slightly. The high-resolution structure of RAdV-1 fibre head is the second known structure of a siadenovirus fibre head domain. The structure shows that the siadenovirus fibre head structure is conserved, but differences in the predicted surface charge suggest that RAdV-1 uses a different natural receptor for cell attachment than TAdV-3. Deletion of the beta-hairpin arm shows little impact on the structure and stability of the siadenovirus fibre heads.
- Departamento de Estructura de Macromoléculas, Centro Nacional de Biotecnología (CNB-CSIC), Calle Darwin 3, E-28049, Madrid, Spain.
Organizational Affiliation: