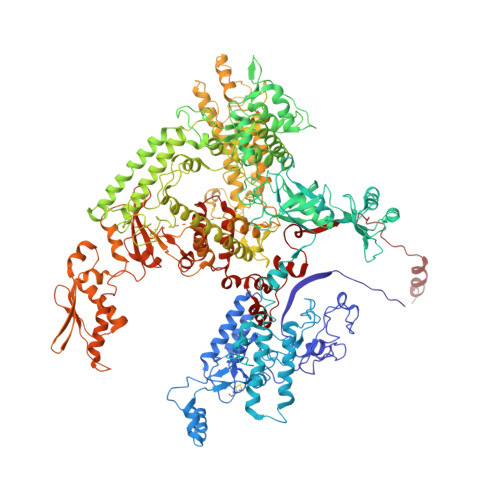
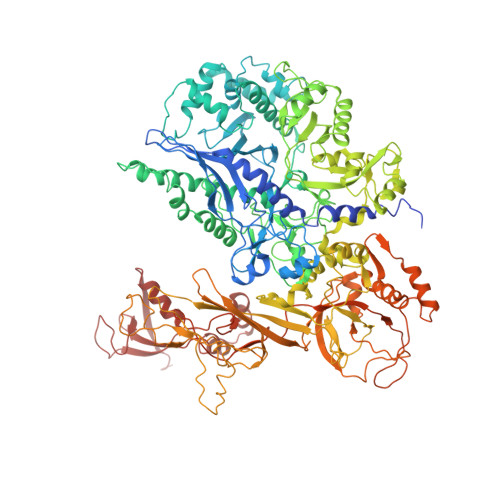
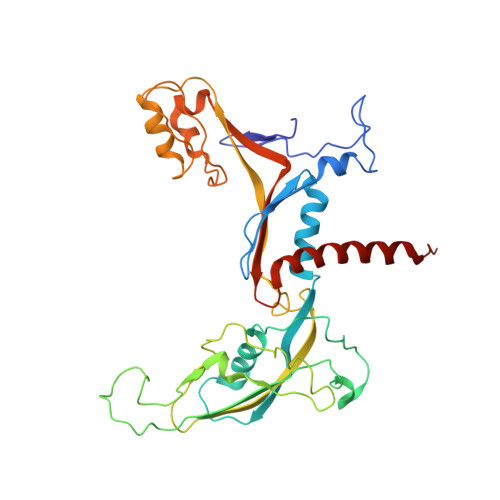
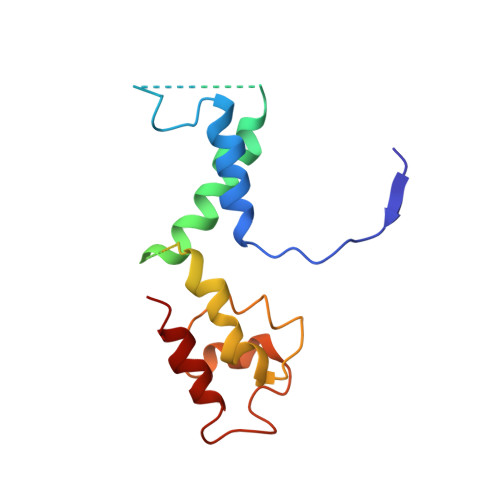
Molecular Structures of Unbound and Transcribing RNA Polymerase III.
Hoffmann, N.A., Jakobi, A.J., Moreno-Morcillo, M., Glatt, S., Kosinski, J., Hagen, W.J.H., Sachse, C., Muller, C.W.(2015) Nature 528: 231
- PubMed: 26605533
- DOI: https://doi.org/10.1038/nature16143
- Primary Citation of Related Structures:
5FJ8, 5FJ9, 5FJA - PubMed Abstract:
Transcription of genes encoding small structured RNAs such as transfer RNAs, spliceosomal U6 small nuclear RNA and ribosomal 5S RNA is carried out by RNA polymerase III (Pol III), the largest yet structurally least characterized eukaryotic RNA polymerase. Here we present the cryo-electron microscopy structures of the Saccharomyces cerevisiae Pol III elongating complex at 3.9 Å resolution and the apo Pol III enzyme in two different conformations at 4.6 and 4.7 Å resolution, respectively, which allow the building of a 17-subunit atomic model of Pol III. The reconstructions reveal the precise orientation of the C82-C34-C31 heterotrimer in close proximity to the stalk. The C53-C37 heterodimer positions residues involved in transcription termination close to the non-template DNA strand. In the apo Pol III structures, the stalk adopts different orientations coupled with closed and open conformations of the clamp. Our results provide novel insights into Pol III-specific transcription and the adaptation of Pol III towards its small transcriptional targets.
- European Molecular Biology Laboratory (EMBL), Structural and Computational Biology Unit, Meyerhofstrasse 1, 69117 Heidelberg, Germany.
Organizational Affiliation: