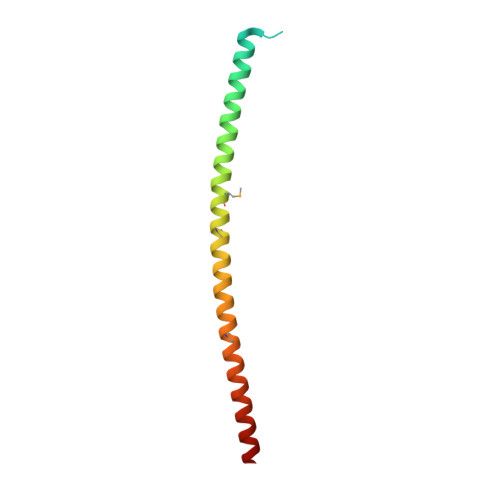Structural basis for the regulatory interactions of proapoptotic Par-4.
Tiruttani Subhramanyam, U.K., Kubicek, J., Eidhoff, U.B., Labahn, J.(2017) Cell Death Differ 24: 1540-1547
- PubMed: 28622290
- DOI: https://doi.org/10.1038/cdd.2017.76
- Primary Citation of Related Structures:
5FIY - PubMed Abstract:
Par-4 is a unique proapoptotic protein with the ability to induce apoptosis selectively in cancer cells. The X-ray crystal structure of the C-terminal domain of Par-4 (Par-4 CC ), which regulates its apoptotic function, was obtained by MAD phasing. Par-4 homodimerizes by forming a parallel coiled-coil structure. The N-terminal half of Par-4 CC contains the homodimerization subdomain. This structure includes a nuclear export signal (Par-4 NES ) sequence, which is masked upon dimerization indicating a potential mechanism for nuclear localization. The heteromeric-interaction models specifically showed that charge interaction is an important factor in the stability of heteromers of the C-terminal leucine zipper subdomain of Par-4 (Par-4 LZ ). These heteromer models also displayed NES masking capacity and therefore the ability to influence intracellular localization.
- Centre for Structural Systems Biology (CSSB), Hamburg, Germany.
Organizational Affiliation: