Darpins as a New Tool for Experimental Phasing in Protein Crystallography
Batyuk, A., Honegger, A., Andres, F., Briand, C., Gruetter, M., Plueckthun, A.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| NI3C DARPIN MUTANT5 HG-SITE N1 | 172 | synthetic construct | Mutation(s): 0 | 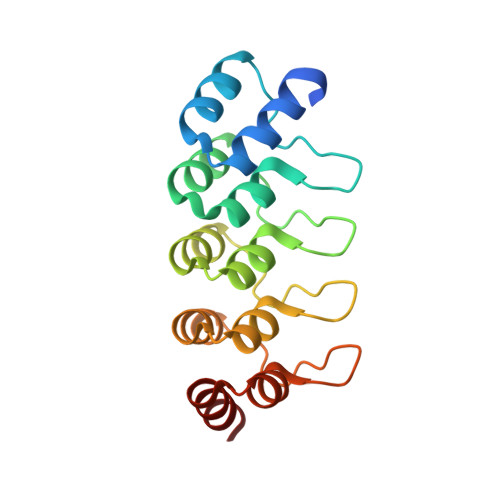 | |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| MALTOSE-BINDING PERIPLASMIC PROTEIN | 395 | Escherichia coli | Mutation(s): 0 | 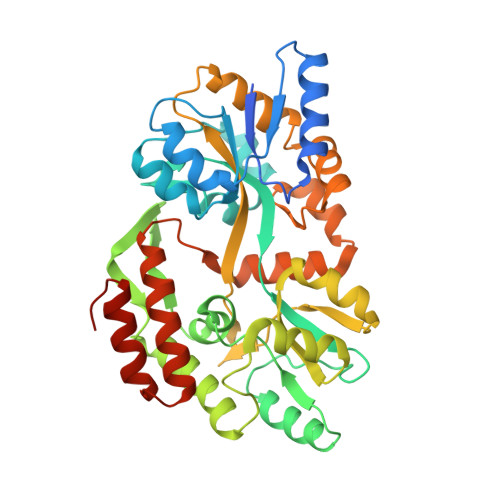 | |
UniProt | |||||
Find proteins for P0AEX9 (Escherichia coli (strain K12)) Explore P0AEX9 Go to UniProtKB: P0AEX9 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P0AEX9 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| HG Query on HG | C [auth A] | MERCURY (II) ION Hg BQPIGGFYSBELGY-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 72.61 | α = 90 |
| b = 43.79 | β = 108.28 |
| c = 82.42 | γ = 90 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| XDS | data reduction |
| XSCALE | data scaling |
| PHASER | phasing |