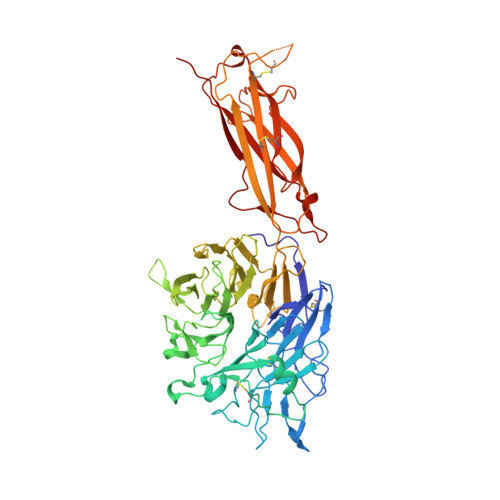
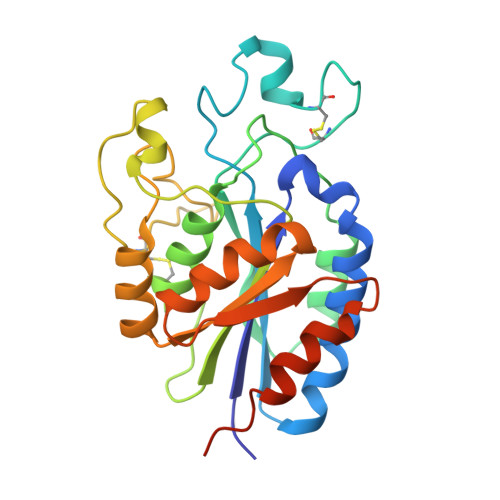
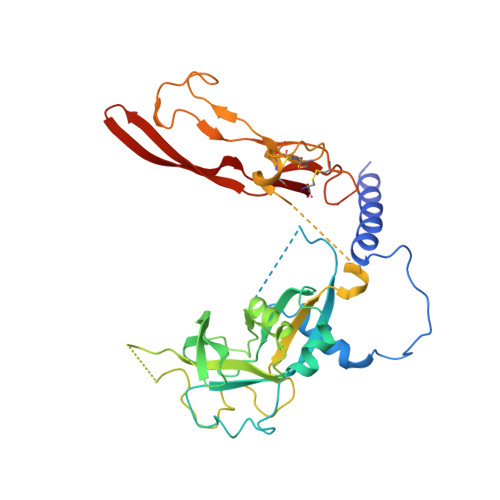
Force interacts with macromolecular structure in activation of TGF-beta.
Dong, X., Zhao, B., Iacob, R.E., Zhu, J., Koksal, A.C., Lu, C., Engen, J.R., Springer, T.A.(2017) Nature 542: 55-59
- PubMed: 28117447
- DOI: https://doi.org/10.1038/nature21035
- Primary Citation of Related Structures:
5FFG, 5FFO - PubMed Abstract:
Integrins are adhesion receptors that transmit force across the plasma membrane between extracellular ligands and the actin cytoskeleton. In activation of the transforming growth factor-β1 precursor (pro-TGF-β1), integrins bind to the prodomain, apply force, and release the TGF-β growth factor. However, we know little about how integrins bind macromolecular ligands in the extracellular matrix or transmit force to them. Here we show how integrin α V β 6 binds pro-TGF-β1 in an orientation biologically relevant for force-dependent release of TGF-β from latency. The conformation of the prodomain integrin-binding motif differs in the presence and absence of integrin binding; differences extend well outside the interface and illustrate how integrins can remodel extracellular matrix. Remodelled residues outside the interface stabilize the integrin-bound conformation, adopt a conformation similar to earlier-evolving family members, and show how macromolecular components outside the binding motif contribute to integrin recognition. Regions in and outside the highly interdigitated interface stabilize a specific integrin/pro-TGF-β orientation that defines the pathway through these macromolecules which actin-cytoskeleton-generated tensile force takes when applied through the integrin β-subunit. Simulations of force-dependent activation of TGF-β demonstrate evolutionary specializations for force application through the TGF-β prodomain and through the β- and not α-subunit of the integrin.
- Children's Hospital Boston and Department of Biological Chemistry and Molecular Pharmacology, Harvard Medical School, Boston, Massachusetts 02115, USA.
Organizational Affiliation: