Unconventional Peptide Presentation by Major Histocompatibility Complex (MHC) Class I Allele HLA-A*02:01: BREAKING CONFINEMENT.
Remesh, S.G., Andreatta, M., Ying, G., Kaever, T., Nielsen, M., McMurtrey, C., Hildebrand, W., Peters, B., Zajonc, D.M.(2017) J Biological Chem 292: 5262-5270
- PubMed: 28179428
- DOI: https://doi.org/10.1074/jbc.M117.776542
- Primary Citation of Related Structures:
5ENW, 5EOT, 5F7D, 5F9J, 5FA3, 5FA4, 5FDW - PubMed Abstract:
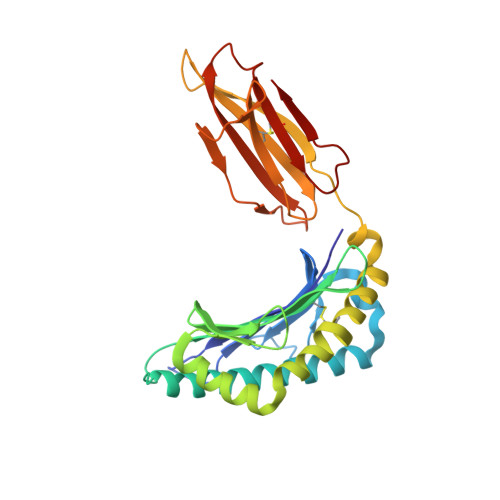
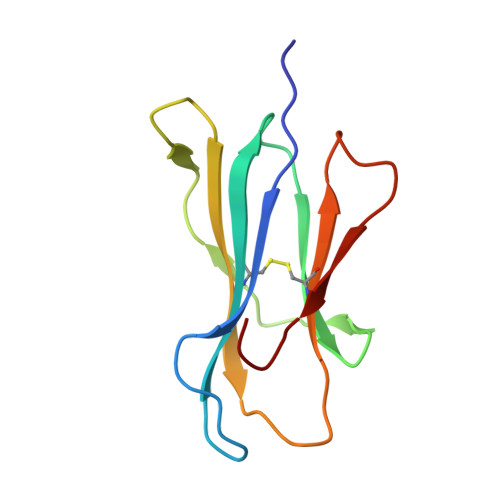
Peptide antigen presentation by major histocompatibility complex (MHC) class I proteins initiates CD8 + T cell-mediated immunity against pathogens and cancers. MHC I molecules typically bind peptides with 9 amino acids in length with both ends tucked inside the major A and F binding pockets. It has been known for a while that longer peptides can also bind by either bulging out of the groove in the middle of the peptide or by binding in a zigzag fashion inside the groove. In a recent study, we identified an alternative binding conformation of naturally occurring peptides from Toxoplasma gondii bound by HLA-A*02:01. These peptides were extended at the C terminus (PΩ) and contained charged amino acids not more than 3 residues after the anchor amino acid at PΩ, which enabled them to open the F pocket and expose their C-terminal extension into the solvent. Here, we show that the mechanism of F pocket opening is dictated by the charge of the first charged amino acid found within the extension. Although positively charged amino acids result in the Tyr-84 swing, amino acids that are negatively charged induce a not previously described Lys-146 lift. Furthermore, we demonstrate that the peptides with alternative binding modes have properties that fit very poorly to the conventional MHC class I pathway and suggest they are presented via alternative means, potentially including cross-presentation via the MHC class II pathway.
- From the Division for Cell Biology and.
Organizational Affiliation: