Structural basis for LIN54 recognition of CHR elements in cell cycle-regulated promoters.
Marceau, A.H., Felthousen, J.G., Goetsch, P.D., Iness, A.N., Lee, H.W., Tripathi, S.M., Strome, S., Litovchick, L., Rubin, S.M.(2016) Nat Commun 7: 12301-12301
- PubMed: 27465258
- DOI: https://doi.org/10.1038/ncomms12301
- Primary Citation of Related Structures:
5FD3 - PubMed Abstract:
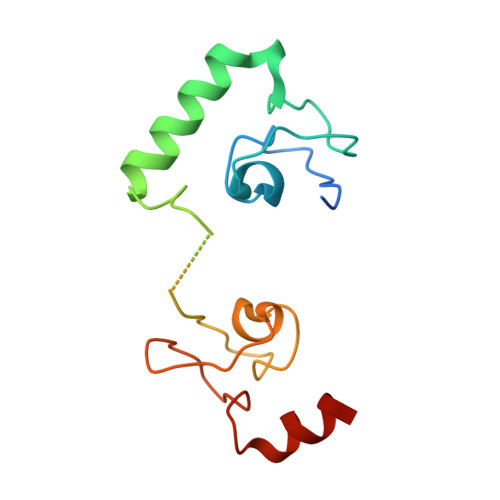
The MuvB complex recruits transcription factors to activate or repress genes with cell cycle-dependent expression patterns. MuvB contains the DNA-binding protein LIN54, which directs the complex to promoter cell cycle genes homology region (CHR) elements. Here we characterize the DNA-binding properties of LIN54 and describe the structural basis for recognition of a CHR sequence. We biochemically define the CHR consensus as TTYRAA and determine that two tandem cysteine rich regions are required for high-affinity DNA association. A crystal structure of the LIN54 DNA-binding domain in complex with a CHR sequence reveals that sequence specificity is conferred by two tyrosine residues, which insert into the minor groove of the DNA duplex. We demonstrate that this unique tyrosine-mediated DNA binding is necessary for MuvB recruitment to target promoters. Our results suggest a model in which MuvB binds near transcription start sites and plays a role in positioning downstream nucleosomes.
- Department of Chemistry and Biochemistry, University of California, 1156 High Street, Santa Cruz, California 95064, USA.
Organizational Affiliation: