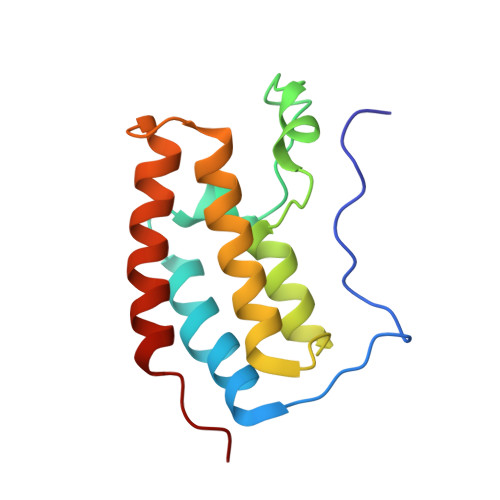
Crystal structure of the first bromodomain of human BRD4 in complex with PNZ5 isoxazole inhibitor
Tallant, C., Clark, P.G.K., Siejka, P., Fedorov, O., Nowak, R., Srikannathasan, V., von Delft, F., Arrowsmith, C.H., Edwards, A.M., Bountra, C., Muller, S., Brennan, P.E., Dixon, D., Knapp, S.To be published.