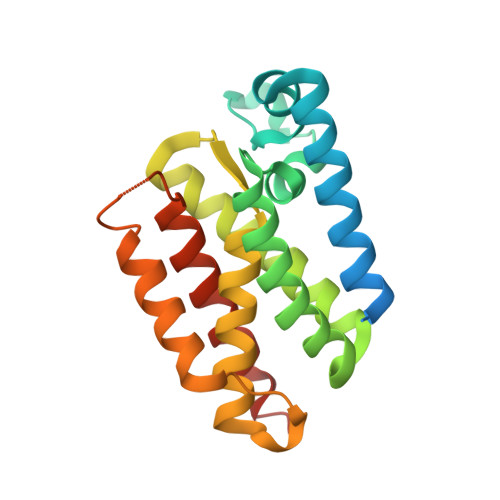
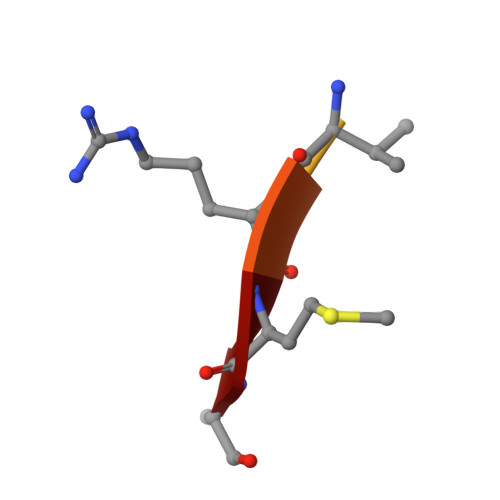
Crystal Structures and Inhibition Kinetics Reveal a Two-Stage Catalytic Mechanism with Drug Design Implications for Rhomboid Proteolysis.
Cho, S., Dickey, S.W., Urban, S.(2016) Mol Cell 61: 329-340
- PubMed: 26805573
- DOI: https://doi.org/10.1016/j.molcel.2015.12.022
- Primary Citation of Related Structures:
5F5B, 5F5D, 5F5G, 5F5J, 5F5K - PubMed Abstract:
Intramembrane proteases signal by releasing proteins from the membrane, but despite their importance, their enzymatic mechanisms remain obscure. We probed rhomboid proteases with reversible, mechanism-based inhibitors that allow precise kinetic analysis and faithfully mimic the transition state structurally. Unexpectedly, inhibition by peptide aldehydes is non-competitive, revealing that in the Michaelis complex, substrate does not contact the catalytic center. Structural analysis in a membrane revealed that all extracellular loops of rhomboid make stabilizing interactions with substrate, but mainly through backbone interactions, explaining rhomboid's broad sequence selectivity. At the catalytic site, the tetrahedral intermediate lies covalently attached to the catalytic serine alone, with the oxyanion stabilized by unusual tripartite interactions with the side chains of H150, N154, and the backbone of S201. We also visualized unexpected substrate-enzyme interactions at the non-essential P2/P3 residues. These "extra" interactions foster potent rhomboid inhibition in living cells, thereby opening avenues for rational design of selective rhomboid inhibitors.
- Howard Hughes Medical Institute, Department of Molecular Biology & Genetics, Johns Hopkins University School of Medicine, Room 507 PCTB, 725 North Wolfe Street, Baltimore, Maryland, USA, 21205.
Organizational Affiliation: