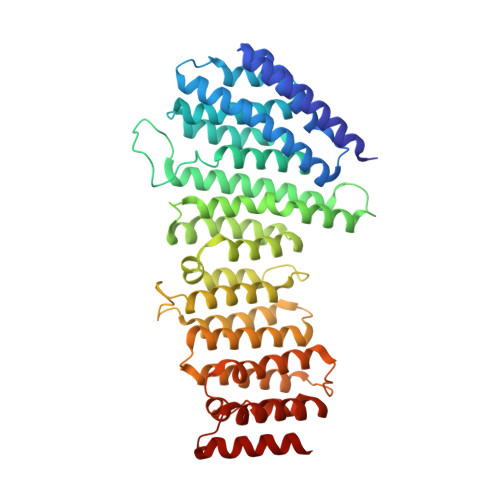
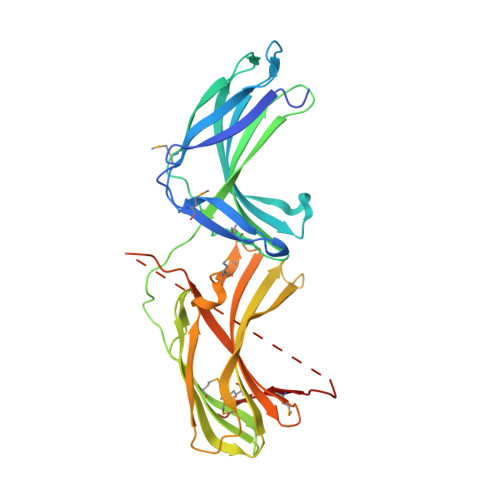
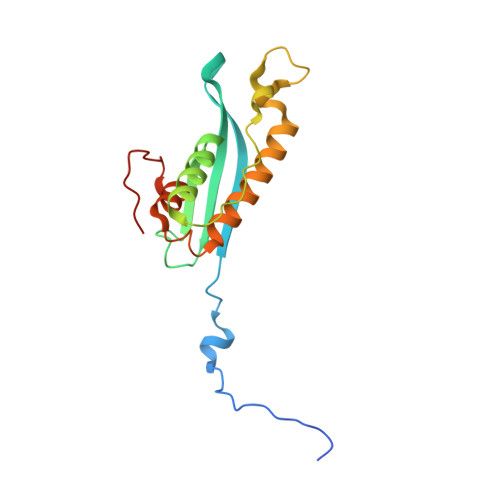
Structural Mechanism for Cargo Recognition by the Retromer Complex.
Lucas, M., Gershlick, D.C., Vidaurrazaga, A., Rojas, A.L., Bonifacino, J.S., Hierro, A.(2016) Cell 167: 1623-1635.e14
- PubMed: 27889239
- DOI: https://doi.org/10.1016/j.cell.2016.10.056
- Primary Citation of Related Structures:
5F0J, 5F0K, 5F0L, 5F0M, 5F0P - PubMed Abstract:
Retromer is a multi-protein complex that recycles transmembrane cargo from endosomes to the trans-Golgi network and the plasma membrane. Defects in retromer impair various cellular processes and underlie some forms of Alzheimer's disease and Parkinson's disease. Although retromer was discovered over 15 years ago, the mechanisms for cargo recognition and recruitment to endosomes have remained elusive. Here, we present an X-ray crystallographic analysis of a four-component complex comprising the VPS26 and VPS35 subunits of retromer, the sorting nexin SNX3, and a recycling signal from the divalent cation transporter DMT1-II. This analysis identifies a binding site for canonical recycling signals at the interface between VPS26 and SNX3. In addition, the structure highlights a network of cooperative interactions among the VPS subunits, SNX3, and cargo that couple signal-recognition to membrane recruitment.
- Structural Biology Unit, CIC bioGUNE, Bizkaia Technology Park, 48160 Derio, Spain.
Organizational Affiliation: