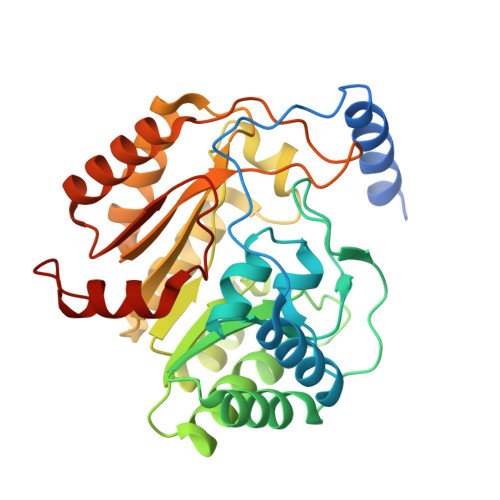
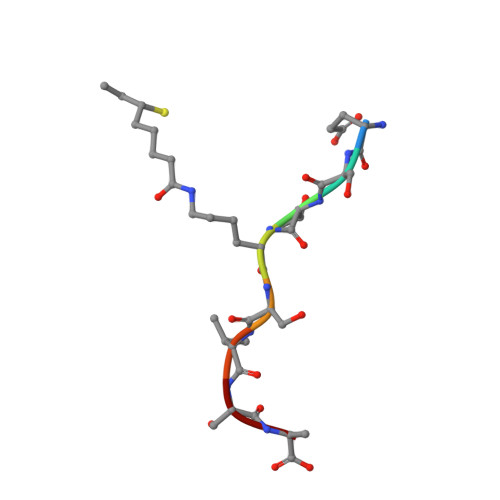
Crystallographic snapshots of sulfur insertion by lipoyl synthase.
McLaughlin, M.I., Lanz, N.D., Goldman, P.J., Lee, K.H., Booker, S.J., Drennan, C.L.(2016) Proc Natl Acad Sci U S A 113: 9446-9450
- PubMed: 27506792
- DOI: https://doi.org/10.1073/pnas.1602486113
- Primary Citation of Related Structures:
5EXI, 5EXJ, 5EXK - PubMed Abstract:
Lipoyl synthase (LipA) catalyzes the insertion of two sulfur atoms at the unactivated C6 and C8 positions of a protein-bound octanoyl chain to produce the lipoyl cofactor. To activate its substrate for sulfur insertion, LipA uses a [4Fe-4S] cluster and S-adenosylmethionine (AdoMet) radical chemistry; the remainder of the reaction mechanism, especially the source of the sulfur, has been less clear. One controversial proposal involves the removal of sulfur from a second (auxiliary) [4Fe-4S] cluster on the enzyme, resulting in destruction of the cluster during each round of catalysis. Here, we present two high-resolution crystal structures of LipA from Mycobacterium tuberculosis: one in its resting state and one at an intermediate state during turnover. In the resting state, an auxiliary [4Fe-4S] cluster has an unusual serine ligation to one of the irons. After reaction with an octanoyllysine-containing 8-mer peptide substrate and 1 eq AdoMet, conditions that allow for the first sulfur insertion but not the second insertion, the serine ligand dissociates from the cluster, the iron ion is lost, and a sulfur atom that is still part of the cluster becomes covalently attached to C6 of the octanoyl substrate. This intermediate structure provides a clear picture of iron-sulfur cluster destruction in action, supporting the role of the auxiliary cluster as the sulfur source in the LipA reaction and describing a radical strategy for sulfur incorporation into completely unactivated substrates.
- Department of Chemistry, Massachusetts Institute of Technology, Cambridge, MA 02139; Department of Chemistry, The Pennsylvania State University, University Park, PA 16802;
Organizational Affiliation: