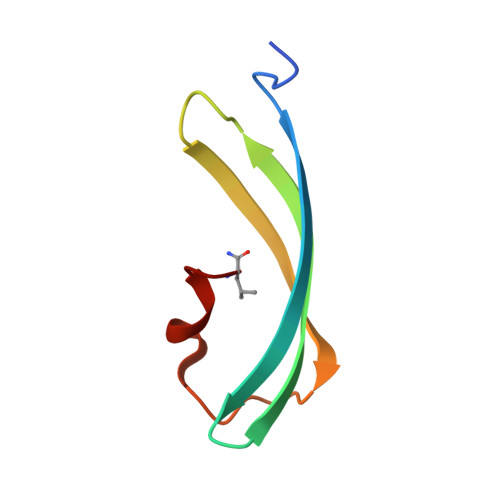
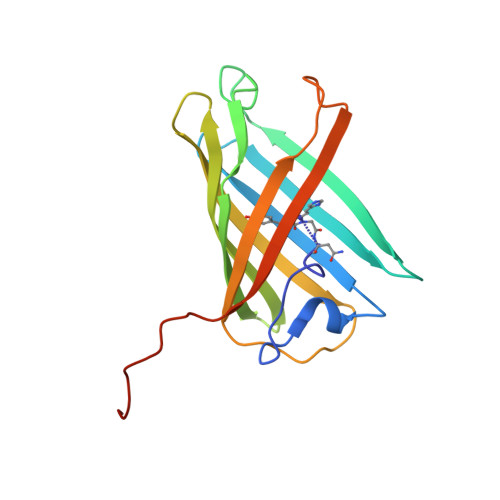
Crystal structure of the fluorescent protein from Dendronephthya sp. in both green and photoconverted red forms.
Pletneva, N.V., Pletnev, S., Pakhomov, A.A., Chertkova, R.V., Martynov, V.I., Muslinkina, L., Dauter, Z., Pletnev, V.Z.(2016) Acta Crystallogr D Struct Biol 72: 922-932
- PubMed: 27487823
- DOI: https://doi.org/10.1107/S205979831601038X
- Primary Citation of Related Structures:
5EXB, 5EXC - PubMed Abstract:
The fluorescent protein from Dendronephthya sp. (DendFP) is a member of the Kaede-like group of photoconvertible fluorescent proteins with a His62-Tyr63-Gly64 chromophore-forming sequence. Upon irradiation with UV and blue light, the fluorescence of DendFP irreversibly changes from green (506 nm) to red (578 nm). The photoconversion is accompanied by cleavage of the peptide backbone at the C(α)-N bond of His62 and the formation of a terminal carboxamide group at the preceding Leu61. The resulting double C(α)=C(β) bond in His62 extends the conjugation of the chromophore π system to include imidazole, providing the red fluorescence. Here, the three-dimensional structures of native green and photoconverted red forms of DendFP determined at 1.81 and 2.14 Å resolution, respectively, are reported. This is the first structure of photoconverted red DendFP to be reported to date. The structure-based mutagenesis of DendFP revealed an important role of positions 142 and 193: replacement of the original Ser142 and His193 caused a moderate red shift in the fluorescence and a considerable increase in the photoconversion rate. It was demonstrated that hydrogen bonding of the chromophore to the Gln116 and Ser105 cluster is crucial for variation of the photoconversion rate. The single replacement Gln116Asn disrupts the hydrogen bonding of Gln116 to the chromophore, resulting in a 30-fold decrease in the photoconversion rate, which was partially restored by a further Ser105Asn replacement.
- Shemyakin-Ovchinnikov Institute of Bioorganic Chemistry, Russian Academy of Sciences, Moscow, Russian Federation.
Organizational Affiliation: