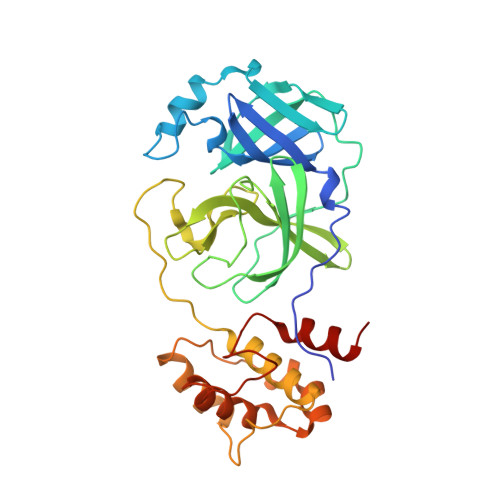
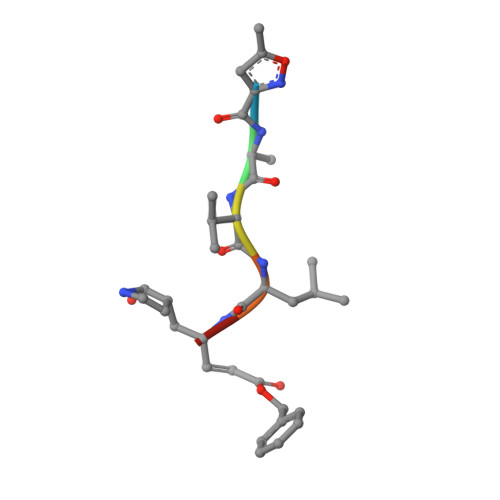
Crystal Structure of Feline Infectious Peritonitis Virus Main Protease in Complex with Synergetic Dual Inhibitors
Wang, F., Chen, C., Liu, X., Yang, K., Xu, X., Yang, H.(2015) J Virol 90: 1910-1917
- PubMed: 26656689
- DOI: https://doi.org/10.1128/JVI.02685-15
- Primary Citation of Related Structures:
5EU8 - PubMed Abstract:
Coronaviruses (CoVs) can cause highly prevalent diseases in humans and animals. Feline infectious peritonitis virus (FIPV) belongs to the genus Alphacoronavirus, resulting in a lethal systemic granulomatous disease called feline infectious peritonitis (FIP), which is one of the most important fatal infectious diseases of cats worldwide. No specific vaccines or drugs have been approved to treat FIP. CoV main proteases (M(pro)s) play a pivotal role in viral transcription and replication, making them an ideal target for drug development. Here, we report the crystal structure of FIPV M(pro) in complex with dual inhibitors, a zinc ion and a Michael acceptor. The complex structure elaborates a unique mechanism of two distinct inhibitors synergizing to inactivate the protease, providing a structural basis to design novel antivirals and suggesting the potential to take advantage of zinc as an adjunct therapy against CoV-associated diseases. Coronaviruses (CoVs) have the largest genome size among all RNA viruses. CoV infection causes various diseases in humans and animals, including severe acute respiratory syndrome (SARS) and Middle East respiratory syndrome (MERS). No approved specific drugs or vaccinations are available to treat their infections. Here, we report a novel dual inhibition mechanism targeting CoV main protease (M(pro)) from feline infectious peritonitis virus (FIPV), which leads to lethal systemic granulomatous disease in cats. M(pro), conserved across all CoV genomes, is essential for viral replication and transcription. We demonstrated that zinc ion and a Michael acceptor-based peptidomimetic inhibitor synergistically inactivate FIPV M(pro). We also solved the structure of FIPV M(pro) complexed with two inhibitors, delineating the structural view of a dual inhibition mechanism. Our study provides new insight into the pharmaceutical strategy against CoV M(pro) through using zinc as an adjuvant therapy to enhance the efficacy of an irreversible peptidomimetic inhibitor.
- School of Life Sciences, Tianjin University, Tianjin, People's Republic of China.
Organizational Affiliation: