Structure of HCV E2 glycoprotein antigenic Epitope II bound to the broadly neutralizing antibody HC84-26
Gao, M., Mariuzza, R.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Anti-HCV E2 Fab HC84-26 heavy chain | 217 | Homo sapiens | Mutation(s): 0 | 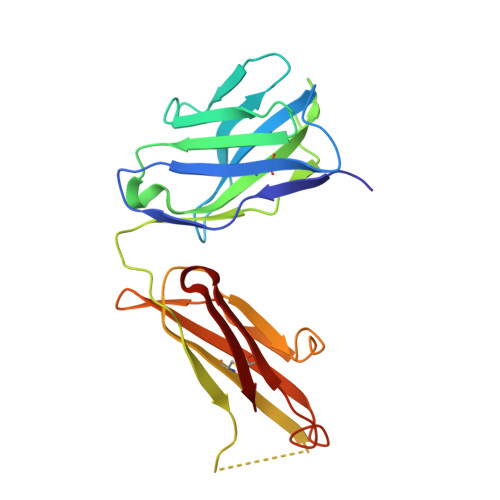 | |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Anti-HCV E2 Fab HC84-26 light chain | 214 | Homo sapiens | Mutation(s): 0 | 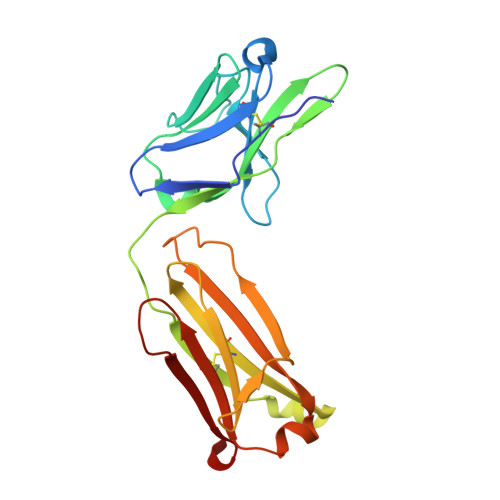 | |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
Find similar proteins by: Sequence | 3D Structure
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| HCV E2 glycoprotein Epitope II | 13 | Hepacivirus hominis | Mutation(s): 0 | 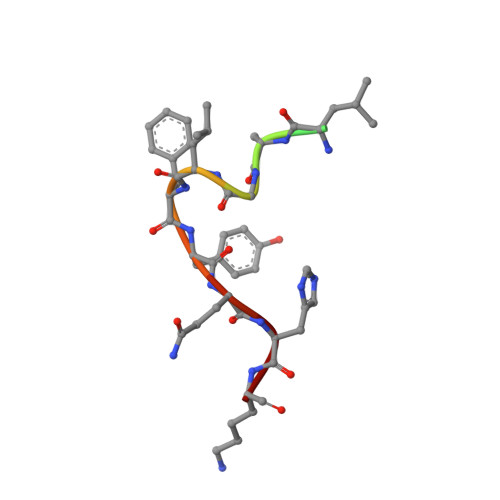 | |
UniProt | |||||
Find proteins for P27958 (Hepatitis C virus genotype 1a (isolate H77)) Explore P27958 Go to UniProtKB: P27958 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P27958 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 37.7 | α = 90 |
| b = 101.24 | β = 90 |
| c = 180.32 | γ = 90 |
| Software Name | Purpose |
|---|---|
| CrystalClear | data collection |
| MOSFLM | data reduction |
| SCALA | data scaling |
| PHASER | phasing |
| PHENIX | refinement |
| Coot | model building |
| PDB_EXTRACT | data extraction |