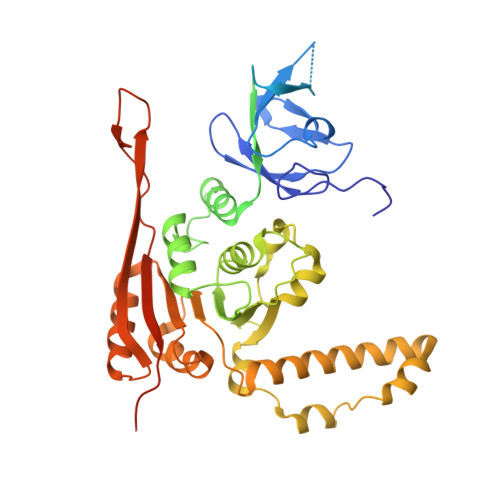
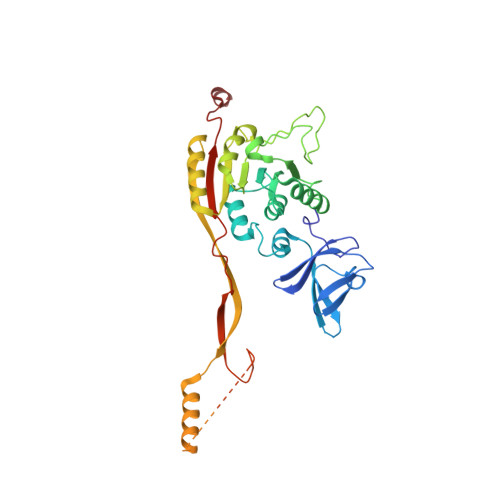
Crystal structure of the two-subunit tRNA m(1)A58 methyltransferase TRM6-TRM61 from Saccharomyces cerevisiae.
Wang, M., Zhu, Y., Wang, C., Fan, X., Jiang, X., Ebrahimi, M., Qiao, Z., Niu, L., Teng, M., Li, X.(2016) Sci Rep 6: 32562-32562
- PubMed: 27582183
- DOI: https://doi.org/10.1038/srep32562
- Primary Citation of Related Structures:
5EQJ, 5ERG - PubMed Abstract:
The N(1) methylation of adenine at position 58 (m(1)A58) of tRNA is an important post-transcriptional modification, which is vital for maintaining the stability of the initiator methionine tRNAi(Met). In eukaryotes, this modification is performed by the TRM6-TRM61 holoenzyme. To understand the molecular mechanism that underlies the cooperation of TRM6 and TRM61 in the methyl transfer reaction, we determined the crystal structure of TRM6-TRM61 holoenzyme from Saccharomyces cerevisiae in the presence and absence of its methyl donor S-Adenosyl-L-methionine (SAM). In the structures, two TRM6-TRM61 heterodimers assemble as a heterotetramer. Both TRM6 and TRM61 subunits comprise an N-terminal β-barrel domain linked to a C-terminal Rossmann-fold domain. TRM61 functions as the catalytic subunit, containing a methyl donor (SAM) binding pocket. TRM6 diverges from TRM61, lacking the conserved motifs used for binding SAM. However, TRM6 cooperates with TRM61 forming an L-shaped tRNA binding regions. Collectively, our results provide a structural basis for better understanding the m(1)A58 modification of tRNA occurred in Saccharomyces cerevisiae.
- Hefei National Laboratory for Physical Sciences at Microscale, Innovation Center for Cell Signalling Network, School of Life Science, University of Science and Technology of China, Hefei, Anhui, 230026, People's Republic of China.
Organizational Affiliation: