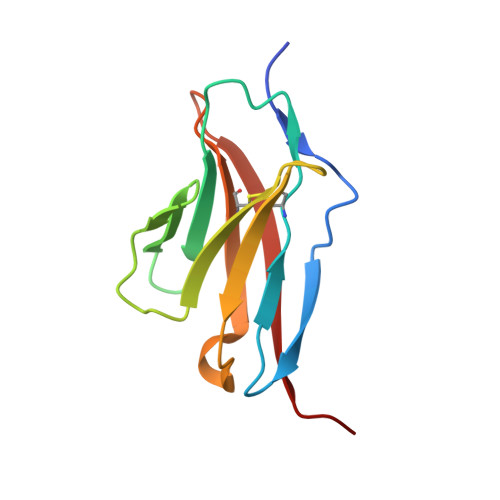
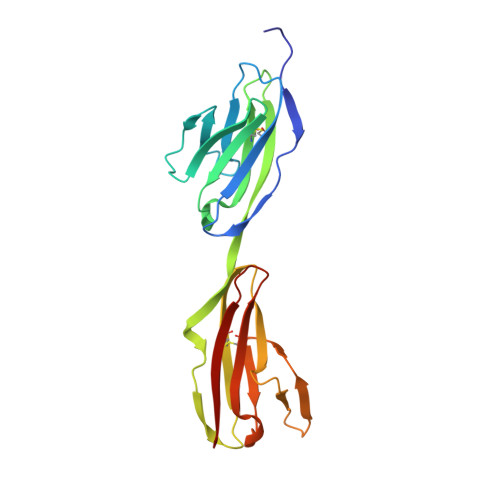
Control of Synaptic Connectivity by a Network of Drosophila IgSF Cell Surface Proteins.
Carrillo, R.A., Ozkan, E., Menon, K.P., Nagarkar-Jaiswal, S., Lee, P.T., Jeon, M., Birnbaum, M.E., Bellen, H.J., Garcia, K.C., Zinn, K.(2015) Cell 163: 1770-1782
- PubMed: 26687361
- DOI: https://doi.org/10.1016/j.cell.2015.11.022
- Primary Citation of Related Structures:
5EO9 - PubMed Abstract:
We have defined a network of interacting Drosophila cell surface proteins in which a 21-member IgSF subfamily, the Dprs, binds to a nine-member subfamily, the DIPs. The structural basis of the Dpr-DIP interaction code appears to be dictated by shape complementarity within the Dpr-DIP binding interface. Each of the six dpr and DIP genes examined here is expressed by a unique subset of larval and pupal neurons. In the neuromuscular system, interactions between Dpr11 and DIP-γ affect presynaptic terminal development, trophic factor responses, and neurotransmission. In the visual system, dpr11 is selectively expressed by R7 photoreceptors that use Rh4 opsin (yR7s). Their primary synaptic targets, Dm8 amacrine neurons, express DIP-γ. In dpr11 or DIP-γ mutants, yR7 terminals extend beyond their normal termination zones in layer M6 of the medulla. DIP-γ is also required for Dm8 survival or differentiation. Our findings suggest that Dpr-DIP interactions are important determinants of synaptic connectivity.
- Division of Biology and Biological Engineering, California Institute of Technology, Pasadena, CA.
Organizational Affiliation: