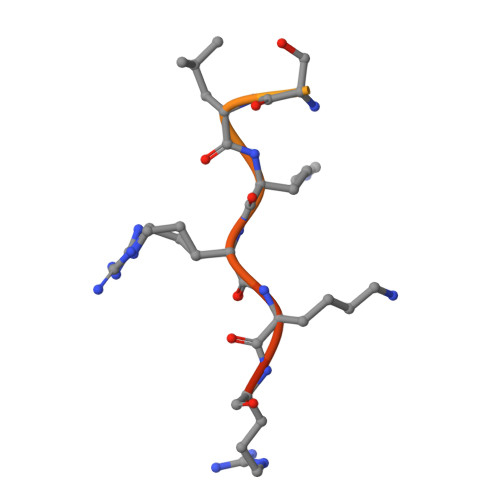
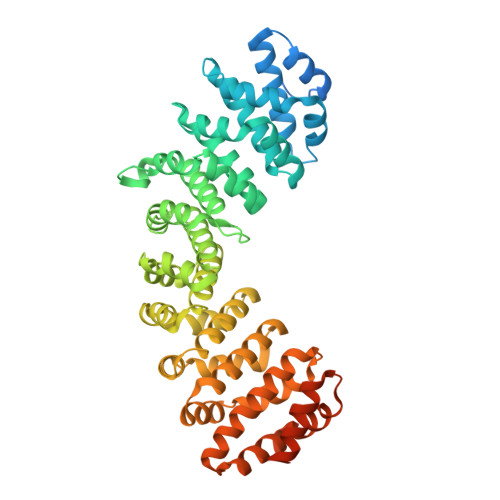
Structural and Calorimetric Studies Demonstrate that Xeroderma Pigmentosum Type G (XPG) Can Be Imported to the Nucleus by a Classical Nuclear Import Pathway via a Monopartite NLS Sequence.
Barros, A.C., Takeda, A.A., Dreyer, T.R., Velazquez-Campoy, A., Kobe, B., Fontes, M.R.(2016) J Mol Biology 428: 2120-2131
- PubMed: 26812207
- DOI: https://doi.org/10.1016/j.jmb.2016.01.019
- Primary Citation of Related Structures:
5EKF, 5EKG - PubMed Abstract:
Xeroderma pigmentosum type G (XPG) proteins are involved in DNA lesion recognition and promotion of nucleotide excision repair. Specific mutations in these proteins may lead to Cockayne syndrome, in which the patients may display severe developmental retardation and neurological abnormalities. No structural information is available for their spacer region or the C-terminal domain, which are important, respectively, for specific nucleotide excision repair activity and substrate specificity, as well as nuclear translocation. Immunofluorescence studies suggested two specific regions of the XPG C-terminus as potential bipartite nuclear localization sequences, which would be responsible for its translocation to the nucleus by the classical nuclear import pathway mediated by the importin-α (Impα). Thus, in order to test these hypotheses and gain insight into the structural basis for the nuclear import process for the XPG protein, we solved the crystal structures of complexes formed by the Impα and peptides corresponding to both putative nuclear localization signal (NLS) sequences (XPG1 and XPG2) and performed isothermal titration calorimetry assays to determine their binding affinities. Structural experiments confirm the binding of both NLS peptides to Impα but, unexpectedly, they bind to the receptor as monopartite NLSs. The isothermal titration calorimetry assays demonstrated that XPG1 and XPG2 peptides bind to two separate binding sites, but with high affinity to the major NLS-binding site of the Impα, resembling classical monopartite SV40 TAg NLS. The results lead to insights about what distinguishes monopartite and bipartite NLSs, as well as the differential roles of XPG1 and XPG2 NLSs in the nuclear localization of XPG.
- Departamento de Física e Biofísica, Instituto de Biociências, Universidade Estadual Paulista, Botucatu, SP, 18618-970 Brazil.
Organizational Affiliation: