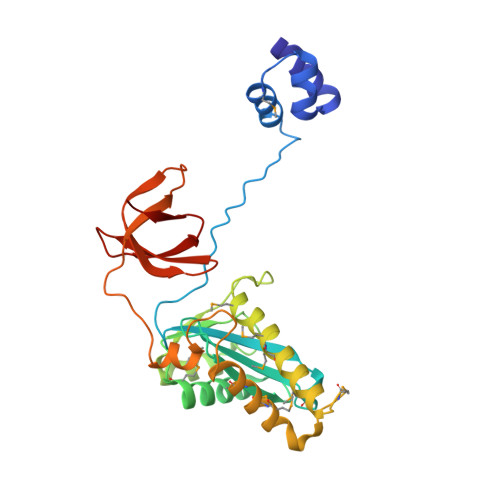
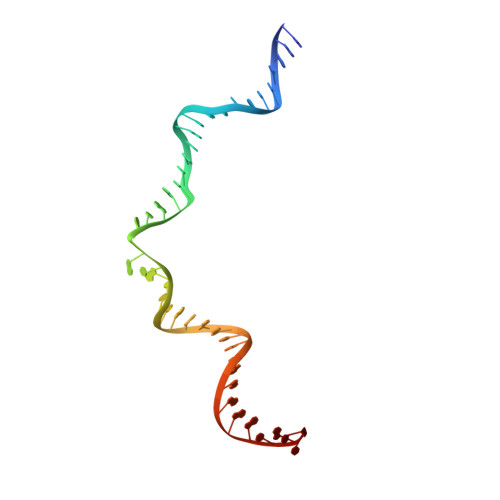
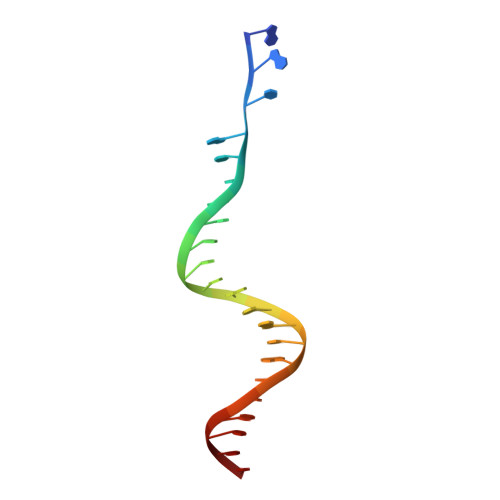
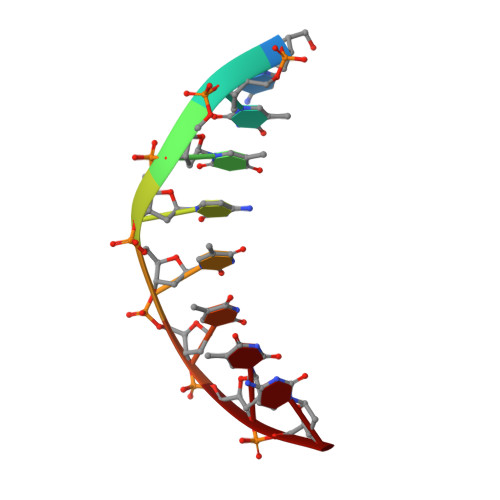
Crystal structure of the Rous sarcoma virus intasome.
Yin, Z., Shi, K., Banerjee, S., Pandey, K.K., Bera, S., Grandgenett, D.P., Aihara, H.(2016) Nature 530: 362-366
- PubMed: 26887497
- DOI: https://doi.org/10.1038/nature16950
- Primary Citation of Related Structures:
5EJK - PubMed Abstract:
Integration of the reverse-transcribed viral DNA into the host genome is an essential step in the life cycle of retroviruses. Retrovirus integrase catalyses insertions of both ends of the linear viral DNA into a host chromosome. Integrase from HIV-1 and closely related retroviruses share the three-domain organization, consisting of a catalytic core domain flanked by amino- and carboxy-terminal domains essential for the concerted integration reaction. Although structures of the tetrameric integrase-DNA complexes have been reported for integrase from prototype foamy virus featuring an additional DNA-binding domain and longer interdomain linkers, the architecture of a canonical three-domain integrase bound to DNA remained elusive. Here we report a crystal structure of the three-domain integrase from Rous sarcoma virus in complex with viral and target DNAs. The structure shows an octameric assembly of integrase, in which a pair of integrase dimers engage viral DNA ends for catalysis while another pair of non-catalytic integrase dimers bridge between the two viral DNA molecules and help capture target DNA. The individual domains of the eight integrase molecules play varying roles to hold the complex together, making an extensive network of protein-DNA and protein-protein contacts that show both conserved and distinct features compared with those observed for prototype foamy virus integrase. Our work highlights the diversity of retrovirus intasome assembly and provides insights into the mechanisms of integration by HIV-1 and related retroviruses.
- Department of Biochemistry, Molecular Biology and Biophysics, University of Minnesota, Minneapolis, Minnesota 55455, USA.
Organizational Affiliation: