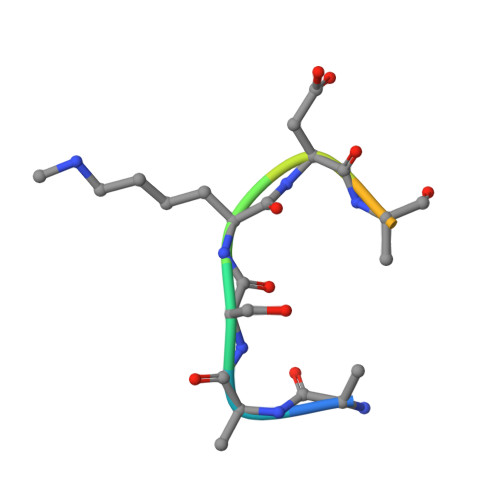
Sulfur-Oxygen Chalcogen Bonding Mediates AdoMet Recognition in the Lysine Methyltransferase SET7/9.
Fick, R.J., Kroner, G.M., Nepal, B., Magnani, R., Horowitz, S., Houtz, R.L., Scheiner, S., Trievel, R.C.(2016) ACS Chem Biol 11: 748-754
- PubMed: 26713889
- DOI: https://doi.org/10.1021/acschembio.5b00852
- Primary Citation of Related Structures:
5EG2 - PubMed Abstract:
Recent studies have demonstrated that carbon-oxygen (CH···O) hydrogen bonds have important roles in S-adenosylmethionine (AdoMet) recognition and catalysis in methyltransferases. Here, we investigate noncovalent interactions that occur between the AdoMet sulfur cation and oxygen atoms in methyltransferase active sites. These interactions represent sulfur-oxygen (S···O) chalcogen bonds in which the oxygen atom donates a lone pair of electrons to the σ antibonding orbital of the AdoMet sulfur atom. Structural, biochemical, and computational analyses of an asparagine mutation in the lysine methyltransferase SET7/9 that abolishes AdoMet S···O chalcogen bonding reveal that this interaction enhances substrate binding affinity relative to the product S-adenosylhomocysteine. Corroborative quantum mechanical calculations demonstrate that sulfonium systems form strong S···O chalcogen bonds relative to their neutral thioether counterparts. An inspection of high-resolution crystal structures reveals the presence of AdoMet S···O chalcogen bonding in different classes of methyltransferases, illustrating that these interactions are not limited to SET domain methyltransferases. Together, these results demonstrate that S···O chalcogen bonds contribute to AdoMet recognition and can enable methyltransferases to distinguish between substrate and product.
- Department of Chemistry and Biochemistry, Utah State University , Logan, Utah 84322, United States.
Organizational Affiliation: