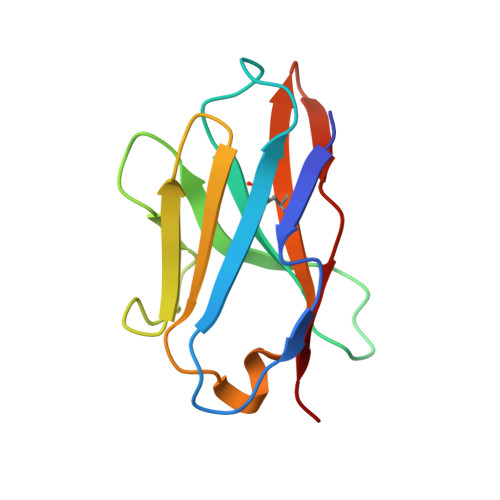
The structural basis of chicken, swine and bovine CD8 alpha alpha dimers provides insight into the co-evolution with MHC I in endotherm species.
Liu, Y., Li, X., Qi, J., Zhang, N., Xia, C.(2016) Sci Rep 6: 24788-24788
- PubMed: 27122108
- DOI: https://doi.org/10.1038/srep24788
- Primary Citation of Related Structures:
5EB9, 5EBG, 5EDX - PubMed Abstract:
It is unclear how the pivotal molecules of the adaptive immune system (AIS) maintain their inherent characteristics and relationships with their co-receptors over the course of co-evolution. CD8α, a fundamental but simple AIS component with only one immunoglobulin variable (IgV) domain, is a good example with which to explore this question because it can fold correctly to form homodimers (CD8αα) and interact with peptide-MHC I (p/MHC I) with low sequence identities between different species. Hereby, we resolved the crystal structures of chicken, swine and bovine CD8αα. They are typical homodimers consisting of two symmetric IgV domains with distinct species specificities. The CD8αα structures indicated that a few highly conserved residues are important in CD8 dimerization and in interacting with p/MHC I. The dimerization of CD8αα mainly depends on the pivotal residues on the dimer interface; in particular, four aromatic residues provide many intermolecular forces and contact areas. Three residues on the surface of CD8α connecting cavities that formed most of the hydrogen bonds with p/MHC I were also completely conserved. Our data propose that a few key conserved residues are able to ensure the CD8α own structural characteristics despite the great sequence variation that occurs during evolution in endotherms.
- Department of Microbiology and Immunology, College of Veterinary Medicine, China Agricultural University, Beijing 100094, China.
Organizational Affiliation: