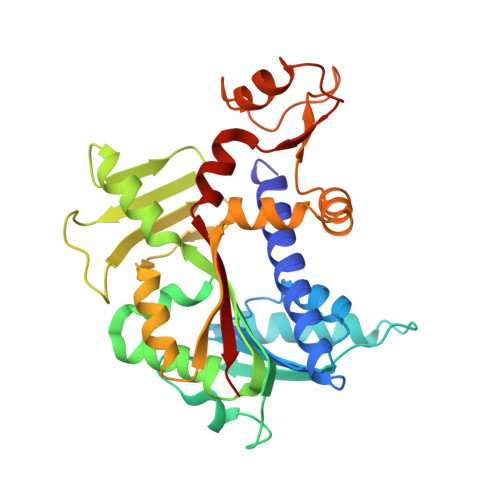
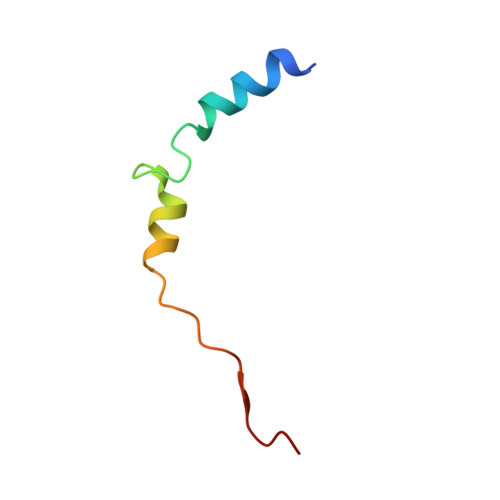
Molecular basis of RNA guanine-7 methyltransferase (RNMT) activation by RAM.
Varshney, D., Petit, A.P., Bueren-Calabuig, J.A., Jansen, C., Fletcher, D.A., Peggie, M., Weidlich, S., Scullion, P., Pisliakov, A.V., Cowling, V.H.(2016) Nucleic Acids Res 44: 10423-10436
- PubMed: 27422871
- DOI: https://doi.org/10.1093/nar/gkw637
- Primary Citation of Related Structures:
5E8J, 5E9J, 5E9W - PubMed Abstract:
Maturation and translation of mRNA in eukaryotes requires the addition of the 7-methylguanosine cap. In vertebrates, the cap methyltransferase, RNA guanine-7 methyltransferase (RNMT), has an activating subunit, RNMT-Activating Miniprotein (RAM). Here we report the first crystal structure of the human RNMT in complex with the activation domain of RAM. A relatively unstructured and negatively charged RAM binds to a positively charged surface groove on RNMT, distal to the active site. This results in stabilisation of a RNMT lobe structure which co-evolved with RAM and is required for RAM binding. Structure-guided mutagenesis and molecular dynamics simulations reveal that RAM stabilises the structure and positioning of the RNMT lobe and the adjacent α-helix hinge, resulting in optimal positioning of helix A which contacts substrates in the active site. Using biophysical and biochemical approaches, we observe that RAM increases the recruitment of the methyl donor, AdoMet (S-adenosyl methionine), to RNMT. Thus we report the mechanism by which RAM allosterically activates RNMT, allowing it to function as a molecular rheostat for mRNA cap methylation.
- Centre for Gene Regulation and Expression, School of Life Sciences, University of Dundee, Dow Street, Dundee DD1 5EH, UK.
Organizational Affiliation: