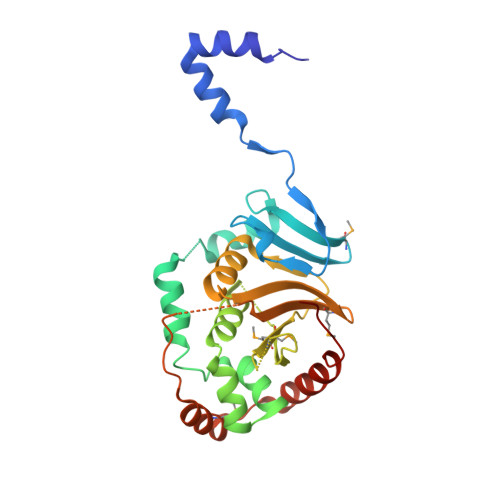
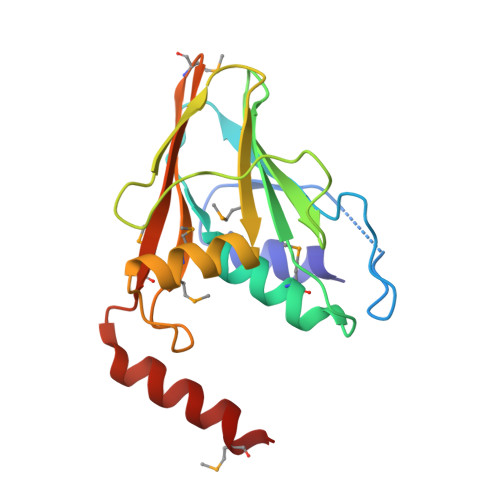
Crystal Structure of the Herpesvirus Nuclear Egress Complex Provides Insights into Inner Nuclear Membrane Remodeling.
Zeev-Ben-Mordehai, T., Weberru, M., Lorenz, M., Cheleski, J., Hellberg, T., Whittle, C., El Omari, K., Vasishtan, D., Dent, K.C., Harlos, K., Franzke, K., Hagen, C., Klupp, B.G., Antonin, W., Mettenleiter, T.C., Grunewald, K.(2015) Cell Rep 13: 2645-2652
- PubMed: 26711332
- DOI: https://doi.org/10.1016/j.celrep.2015.11.008
- Primary Citation of Related Structures:
5E8C, 5FKI - PubMed Abstract:
Although nucleo-cytoplasmic transport is typically mediated through nuclear pore complexes, herpesvirus capsids exit the nucleus via a unique vesicular pathway. Together, the conserved herpesvirus proteins pUL31 and pUL34 form the heterodimeric nuclear egress complex (NEC), which, in turn, mediates the formation of tight-fitting membrane vesicles around capsids at the inner nuclear membrane. Here, we present the crystal structure of the pseudorabies virus NEC. The structure revealed that a zinc finger motif in pUL31 and an extensive interaction network between the two proteins stabilize the complex. Comprehensive mutational analyses, characterized both in situ and in vitro, indicated that the interaction network is not redundant but rather complementary. Fitting of the NEC crystal structure into the recently determined cryoEM-derived hexagonal lattice, formed in situ by pUL31 and pUL34, provided details on the molecular basis of NEC coat formation and inner nuclear membrane remodeling.
- Division of Structural Biology, Wellcome Trust Centre for Human Genetics, University of Oxford, Roosevelt Drive, Oxford OX3 7BN, UK.
Organizational Affiliation: