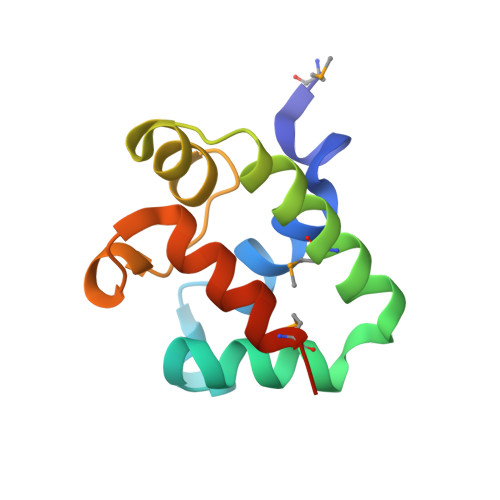
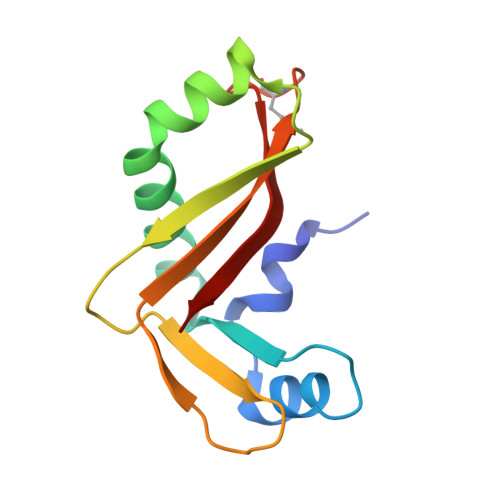
The CDI toxin of Yersinia kristensenii is a novel bacterial member of the RNase A superfamily.
Batot, G., Michalska, K., Ekberg, G., Irimpan, E.M., Joachimiak, G., Jedrzejczak, R., Babnigg, G., Hayes, C.S., Joachimiak, A., Goulding, C.W.(2017) Nucleic Acids Res 45: 5013-5025
- PubMed: 28398546
- DOI: https://doi.org/10.1093/nar/gkx230
- Primary Citation of Related Structures:
5E3E - PubMed Abstract:
Contact-dependent growth inhibition (CDI) is an important mechanism of inter-bacterial competition found in many Gram-negative pathogens. CDI+ cells express cell-surface CdiA proteins that bind neighboring bacteria and deliver C-terminal toxin domains (CdiA-CT) to inhibit target-cell growth. CDI+ bacteria also produce CdiI immunity proteins, which specifically neutralize cognate CdiA-CT toxins to prevent self-inhibition. Here, we present the crystal structure of the CdiA-CT/CdiIYkris complex from Yersinia kristensenii ATCC 33638. CdiA-CTYkris adopts the same fold as angiogenin and other RNase A paralogs, but the toxin does not share sequence similarity with these nucleases and lacks the characteristic disulfide bonds of the superfamily. Consistent with the structural homology, CdiA-CTYkris has potent RNase activity in vitro and in vivo. Structure-guided mutagenesis reveals that His175, Arg186, Thr276 and Tyr278 contribute to CdiA-CTYkris activity, suggesting that these residues participate in substrate binding and/or catalysis. CdiIYkris binds directly over the putative active site and likely neutralizes toxicity by blocking access to RNA substrates. Significantly, CdiA-CTYkris is the first non-vertebrate protein found to possess the RNase A superfamily fold, and homologs of this toxin are associated with secretion systems in many Gram-negative and Gram-positive bacteria. These observations suggest that RNase A-like toxins are commonly deployed in inter-bacterial competition.
- Department of Molecular Biology & Biochemistry, University of California Irvine, Irvine, CA 92697, USA.
Organizational Affiliation: