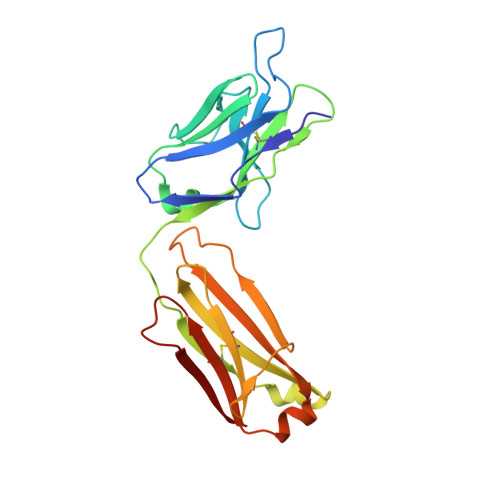
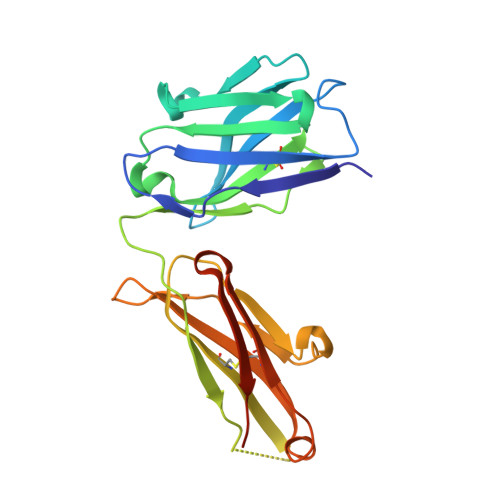
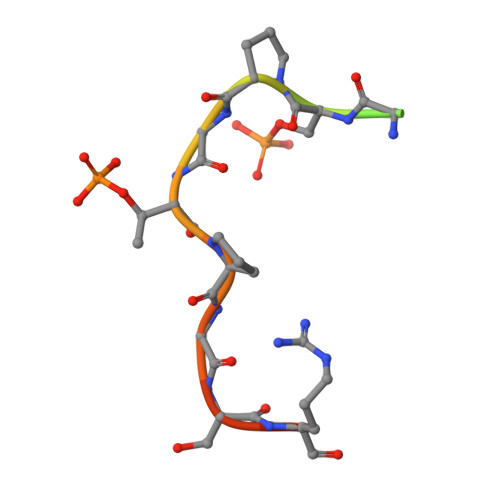
Epitope mapping and structural basis for the recognition of phosphorylated tau by the anti-tau antibody AT8.
Malia, T.J., Teplyakov, A., Ernst, R., Wu, S.J., Lacy, E.R., Liu, X., Vandermeeren, M., Mercken, M., Luo, J., Sweet, R.W., Gilliland, G.L.(2016) Proteins 84: 427-434
- PubMed: 26800003
- DOI: https://doi.org/10.1002/prot.24988
- Primary Citation of Related Structures:
5E2T, 5E2U, 5E2V, 5E2W - PubMed Abstract:
Microtubule-associated protein tau becomes abnormally phosphorylated in Alzheimer's disease and other tauopathies and forms aggregates of paired helical filaments (PHF-tau). AT8 is a PHF-tau-specific monoclonal antibody that is a commonly used marker of neuropathology because of its recognition of abnormally phosphorylated tau. Previous reports described the AT8 epitope to include pS202/pT205. Our studies support and extend previous findings by also identifying pS208 as part of the binding epitope. We characterized the phosphoepitope of AT8 through both peptide binding studies and costructures with phosphopeptides. From the cocrystal structure of AT8 Fab with the diphosphorylated (pS202/pT205) peptide, it appeared that an additional phosphorylation at S208 would also be accommodated by AT8. Phosphopeptide binding studies showed that AT8 bound to the triply phosphorylated tau peptide (pS202/pT205/pS208) 30-fold stronger than to the pS202/pT205 peptide, supporting the role of pS208 in AT8 recognition. We also show that the binding kinetics of the triply phosphorylated peptide pS202/pT205/pS208 was remarkably similar to that of PHF-tau. The costructure of AT8 Fab with a pS202/pT205/pS208 peptide shows that the interaction interface involves all six CDRs and tau residues 202-209. All three phosphorylation sites are recognized by AT8, with pT205 acting as the anchor. Crystallization of the Fab/peptide complex under acidic conditions shows that CDR-L2 is prone to unfolding and precludes peptide binding, and may suggest a general instability in the antibody.
- Janssen Research & Development, LLC, 1400 McKean Road, Spring House, Pennsylvania, 19477.
Organizational Affiliation: