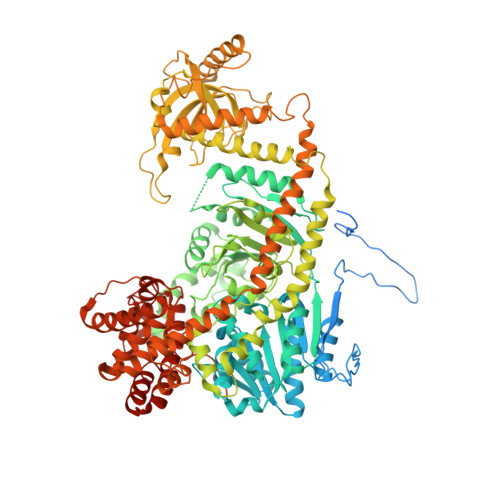
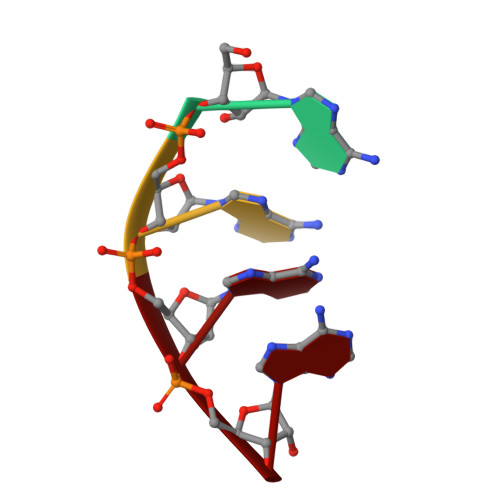
Structure of the frequency-interacting RNA helicase: a protein interaction hub for the circadian clock.
Conrad, K.S., Hurley, J.M., Widom, J., Ringelberg, C.S., Loros, J.J., Dunlap, J.C., Crane, B.R.(2016) EMBO J 35: 1707-1719
- PubMed: 27340124
- DOI: https://doi.org/10.15252/embj.201694327
- Primary Citation of Related Structures:
4XGT, 5DZR, 5E02 - PubMed Abstract:
In the Neurospora crassa circadian clock, a protein complex of frequency (FRQ), casein kinase 1a (CK1a), and the FRQ-interacting RNA Helicase (FRH) rhythmically represses gene expression by the white-collar complex (WCC). FRH crystal structures in several conformations and bound to ADP/RNA reveal differences between FRH and the yeast homolog Mtr4 that clarify the distinct role of FRH in the clock. The FRQ-interacting region at the FRH N-terminus has variable structure in the absence of FRQ A known mutation that disrupts circadian rhythms (R806H) resides in a positively charged surface of the KOW domain, far removed from the helicase core. We show that changes to other similarly located residues modulate interactions with the WCC and FRQ A V142G substitution near the N-terminus also alters FRQ and WCC binding to FRH, but produces an unusual short clock period. These data support the assertion that FRH helicase activity does not play an essential role in the clock, but rather FRH acts to mediate contacts among FRQ, CK1a and the WCC through interactions involving its N-terminus and KOW module.
- Department of Chemistry and Chemical Biology, Cornell University, Ithaca, NY, USA.
Organizational Affiliation: