The Rational Design of Selective Benzoxazepin Inhibitors of the alpha-Isoform of Phosphoinositide 3-Kinase Culminating in the Identification of (S)-2-((2-(1-Isopropyl-1H-1,2,4-triazol-5-yl)-5,6-dihydrobenzo[f]imidazo[1,2-d][1,4]oxazepin-9-yl)oxy)propanamide (GDC-0326).
Heffron, T.P., Heald, R.A., Ndubaku, C., Wei, B., Augistin, M., Do, S., Edgar, K., Eigenbrot, C., Friedman, L., Gancia, E., Jackson, P.S., Jones, G., Kolesnikov, A., Lee, L.B., Lesnick, J.D., Lewis, C., McLean, N., Mortl, M., Nonomiya, J., Pang, J., Price, S., Prior, W.W., Salphati, L., Sideris, S., Staben, S.T., Steinbacher, S., Tsui, V., Wallin, J., Sampath, D., Olivero, A.G.(2016) J Med Chem 59: 985-1002
- PubMed: 26741947
- DOI: https://doi.org/10.1021/acs.jmedchem.5b01483
- Primary Citation of Related Structures:
5DXH, 5DXT, 5DXU - PubMed Abstract:
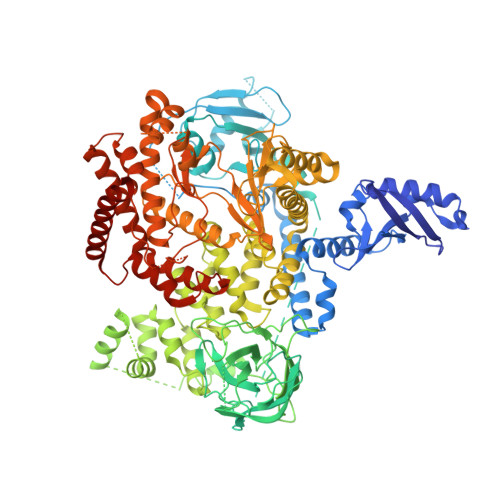
Inhibitors of the class I phosphoinositide 3-kinase (PI3K) isoform PI3Kα have received substantial attention for their potential use in cancer therapy. Despite the particular attraction of targeting PI3Kα, achieving selectivity for the inhibition of this isoform has proved challenging. Herein we report the discovery of inhibitors of PI3Kα that have selectivity over the other class I isoforms and all other kinases tested. In GDC-0032 (3, taselisib), we previously minimized inhibition of PI3Kβ relative to the other class I insoforms. Subsequently, we extended our efforts to identify PI3Kα-specific inhibitors using PI3Kα crystal structures to inform the design of benzoxazepin inhibitors with selectivity for PI3Kα through interactions with a nonconserved residue. Several molecules selective for PI3Kα relative to the other class I isoforms, as well as other kinases, were identified. Optimization of properties related to drug metabolism then culminated in the identification of the clinical candidate GDC-0326 (4).
- Genentech, Inc. , 1 DNA Way, South San Francisco, California 94080, United States.
Organizational Affiliation: