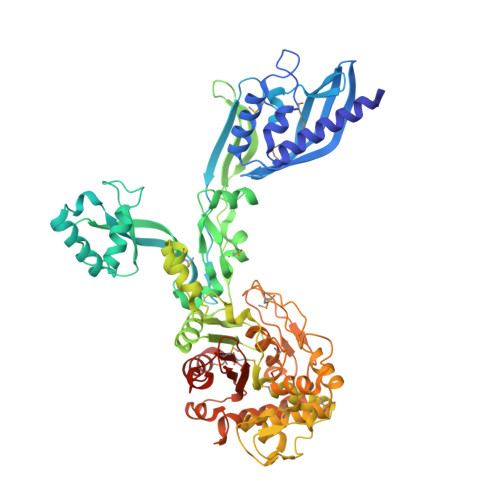
2.95 Angstrom Crystal Structure of the Dimeric Form of Penicillin Binding Protein 2 Prime from Enterococcus faecium.
Minasov, G., Wawrzak, Z., Shuvalova, L., Dubrovska, I., Flores, K., Filippova, E., Grimshaw, S., Kwon, K., Anderson, W.F., Center for Structural Genomics of Infectious Diseases (CSGID)To be published.