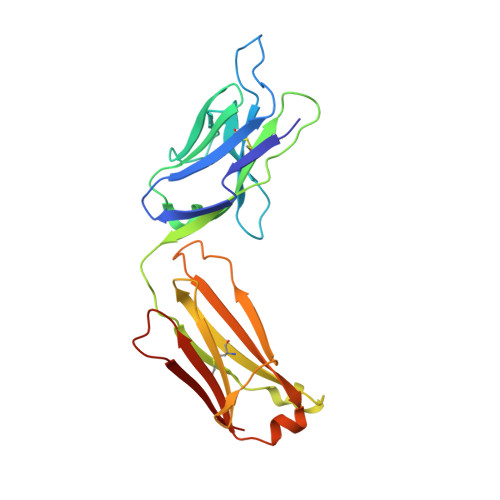
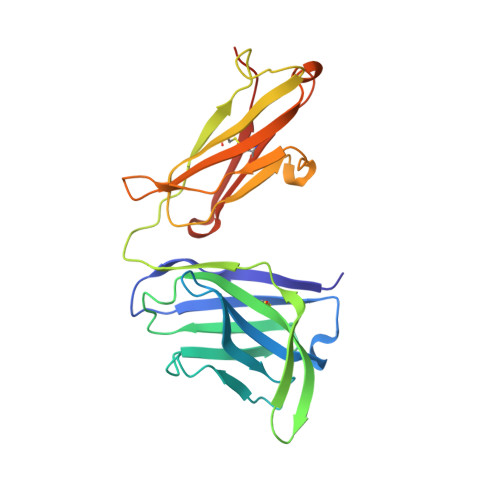
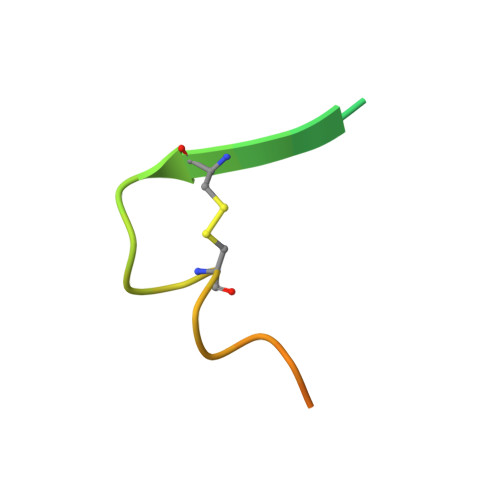
Molecular basis for epitope recognition by non-neutralizing anti-gp41 antibody F240.
Gohain, N., Tolbert, W.D., Orlandi, C., Richard, J., Ding, S., Chen, X., Bonsor, D.A., Sundberg, E.J., Lu, W., Ray, K., Finzi, A., Lewis, G.K., Pazgier, M.(2016) Sci Rep 6: 36685-36685
- PubMed: 27827447
- DOI: https://doi.org/10.1038/srep36685
- Primary Citation of Related Structures:
5DRZ - PubMed Abstract:
Antibody-dependent cell-mediated cytotoxicity (ADCC) by non-neutralizing antibodies (nnAbs) specific to the HIV envelope (Env) glycoproteins present at the surface of virus sensitized or infected cells plays a role in the effective adaptive immune response to HIV. Here, we explore the molecular basis for the epitope at the disulfide loop region (DLR) of the principal immunodominant domain of gp41, recognized by the well-known nnAb F240. Our structural studies reveal details of the F240-gp41 interface and describe a structure of DLR that is distinct from known conformations of this region studied in the context of either CD4-unliganded Env trimer or the gp41 peptide in the unbound state. These data coupled with binding and functional analyses indicate that F240 recognizes non-trimeric Env forms which are significantly overexpressed on intact virions but poorly represented at surfaces of cells infected with infectious molecular clones and endogenously-infected CD4 T cells from HIV-1-infected individuals. Furthermore, although we detect ADCC activities of F240 against cells spinoculated with intact virions, our data suggest that these activities result from F240 recognition of gp41 stumps or misfolded Env variants present on virions rather than its ability to recognize functional gp41 transition structures emerging on trimeric Env post CD4 receptor engagement.
- Division of Vaccine Research of Institute of Human Virology, the University of Maryland School of Medicine, Baltimore, USA.
Organizational Affiliation: