Generation and Affinity Maturation of Neutralizing Anti-Sclerostin Antibodies
Boschert, V., Frisch, C., Back, J.W., van Pee, K., Weidauer, S.E., Muth, E.-M., Schmieder, P., Beerbaum, M., Knappik, A., Timmerman, P., Mueller, T.D.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| AbD09097 Fab heavy chain | A [auth H] | 248 | Homo sapiens | Mutation(s): 0 | 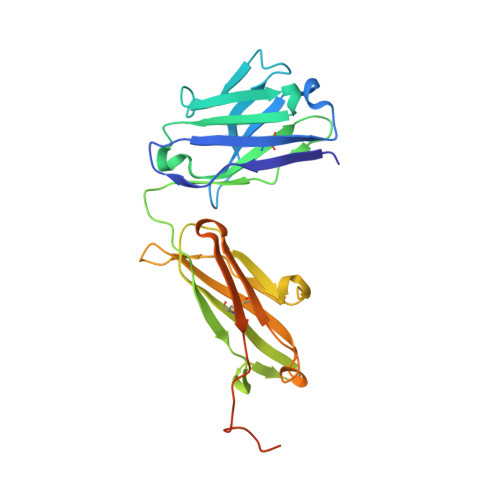 |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| AbD09097 Fab light chain | B [auth L] | 215 | Homo sapiens | Mutation(s): 0 | 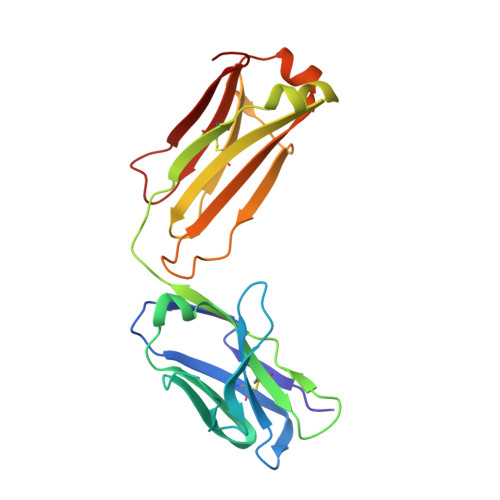 |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 45.193 | α = 90 |
| b = 78.495 | β = 95.71 |
| c = 59.196 | γ = 90 |
| Software Name | Purpose |
|---|---|
| CrystalClear | data collection |
| CrystalClear | data reduction |
| CrystalClear | data scaling |
| PHASER | phasing |
| REFMAC | refinement |
| PDB_EXTRACT | data extraction |
| Funding Organization | Location | Grant Number |
|---|---|---|
| German Research Foundation | Germany | DFG MU1095/5-1 |
| European Union | TALOS, HEALTH-F2-2008-201099 |