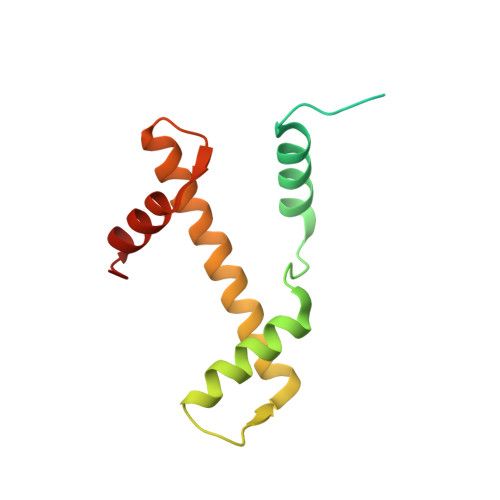
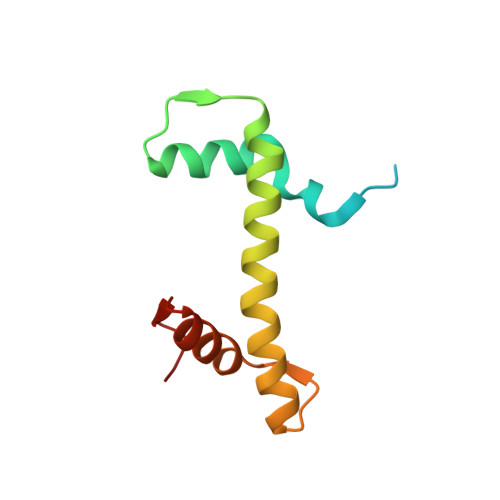
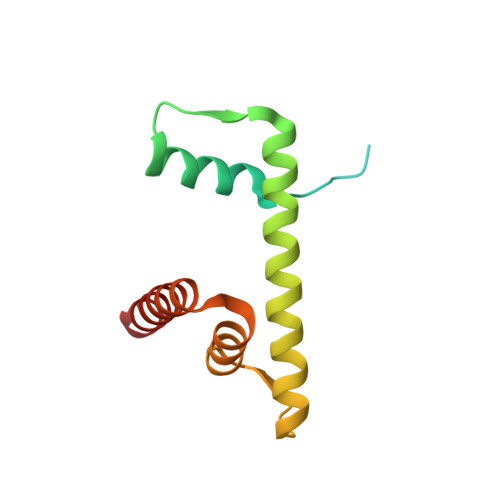
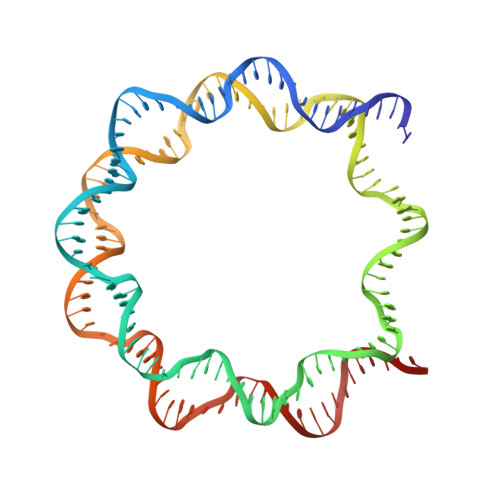
Allosteric cross-talk in chromatin can mediate drug-drug synergy
Adhireksan, Z., Palermo, G., Riedel, T., Ma, Z., Muhammad, R., Rothlisberger, U., Dyson, P.J., Davey, C.A.(2017) Nat Commun 8: 14860-14860
- PubMed: 28358030
- DOI: https://doi.org/10.1038/ncomms14860
- Primary Citation of Related Structures:
5DNM, 5DNN - PubMed Abstract:
Exploitation of drug-drug synergism and allostery could yield superior therapies by capitalizing on the immensely diverse, but highly specific, potential associated with the biological macromolecular landscape. Here we describe a drug-drug synergy mediated by allosteric cross-talk in chromatin, whereby the binding of one drug alters the activity of the second. We found two unrelated drugs, RAPTA-T and auranofin, that yield a synergistic activity in killing cancer cells, which coincides with a substantially greater number of chromatin adducts formed by one of the compounds when adducts from the other agent are also present. We show that this occurs through an allosteric mechanism within the nucleosome, whereby defined histone adducts of one drug promote reaction of the other drug at a distant, specific histone site. This opens up possibilities for epigenetic targeting and suggests that allosteric modulation in nucleosomes may have biological relevance and potential for therapeutic interventions.
- School of Biological Sciences, Nanyang Technological University, 60 Nanyang Drive, Singapore 637551, Singapore.
Organizational Affiliation: