Structure of ATP synthase from Paracoccus denitrificans determined by X-ray crystallography at 4.0 angstrom resolution.
Morales-Rios, E., Montgomery, M.G., Leslie, A.G., Walker, J.E.(2015) Proc Natl Acad Sci U S A 112: 13231-13236
- PubMed: 26460036
- DOI: https://doi.org/10.1073/pnas.1517542112
- Primary Citation of Related Structures:
5DN6 - PubMed Abstract:
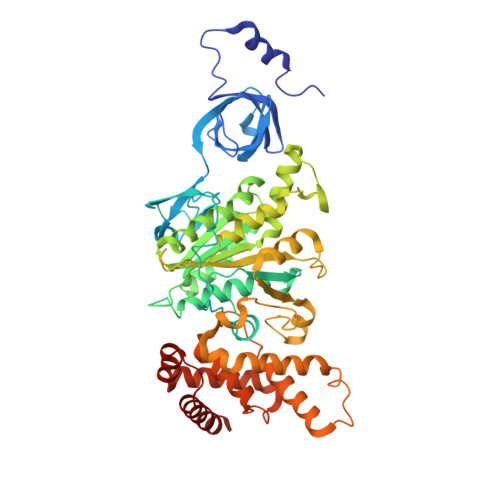
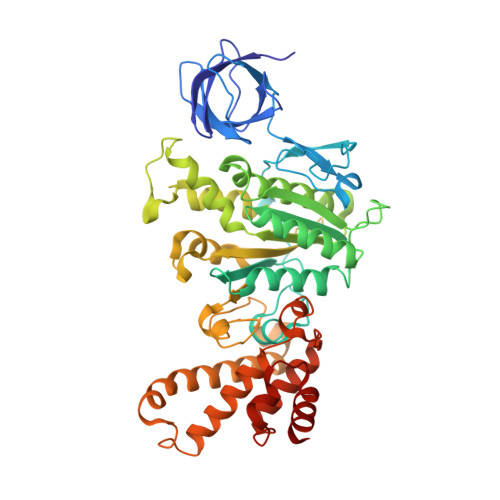
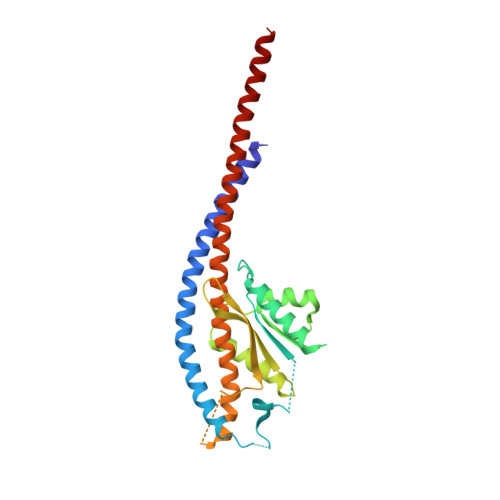
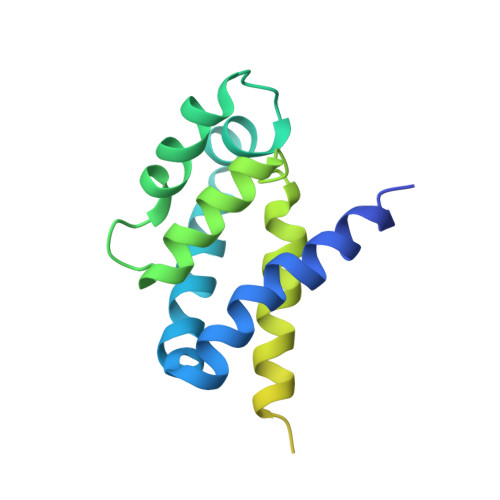
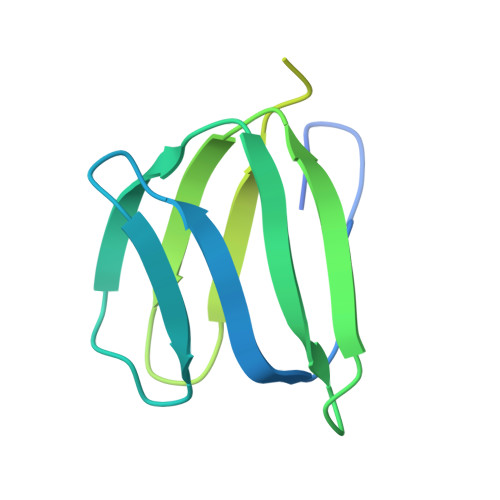
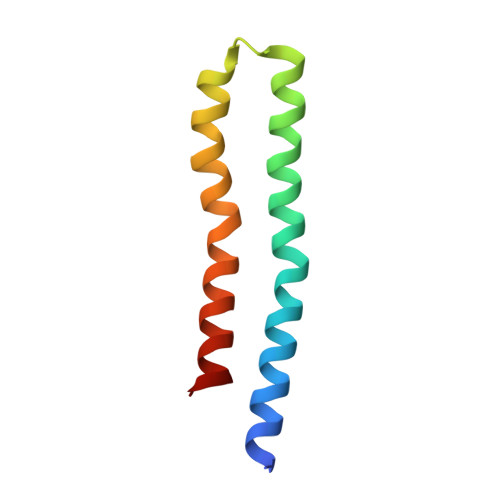
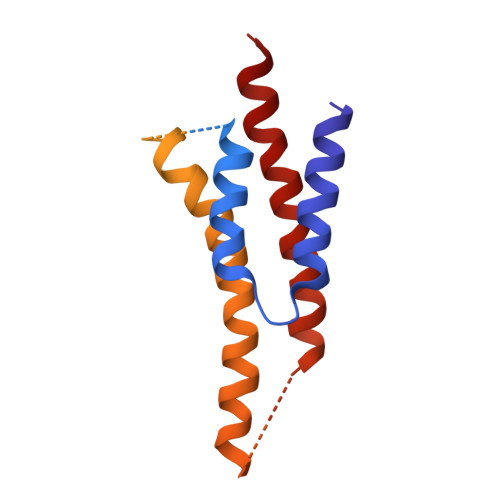
The structure of the intact ATP synthase from the α-proteobacterium Paracoccus denitrificans, inhibited by its natural regulatory ζ-protein, has been solved by X-ray crystallography at 4.0 Å resolution. The ζ-protein is bound via its N-terminal α-helix in a catalytic interface in the F1 domain. The bacterial F1 domain is attached to the membrane domain by peripheral and central stalks. The δ-subunit component of the peripheral stalk binds to the N-terminal regions of two α-subunits. The stalk extends via two parallel long α-helices, one in each of the related b and b' subunits, down a noncatalytic interface of the F1 domain and interacts in an unspecified way with the a-subunit in the membrane domain. The a-subunit lies close to a ring of 12 c-subunits attached to the central stalk in the F1 domain, and, together, the central stalk and c-ring form the enzyme's rotor. Rotation is driven by the transmembrane proton-motive force, by a mechanism where protons pass through the interface between the a-subunit and c-ring via two half-channels in the a-subunit. These half-channels are probably located in a bundle of four α-helices in the a-subunit that are tilted at ∼30° to the plane of the membrane. Conserved polar residues in the two α-helices closest to the c-ring probably line the proton inlet path to an essential carboxyl group in the c-subunit in the proton uptake site and a proton exit path from the proton release site. The structure has provided deep insights into the workings of this extraordinary molecular machine.
- Medical Research Council Mitochondrial Biology Unit, Cambridge Biomedical Campus, Cambridge CB2 0XY, United Kingdom;
Organizational Affiliation: