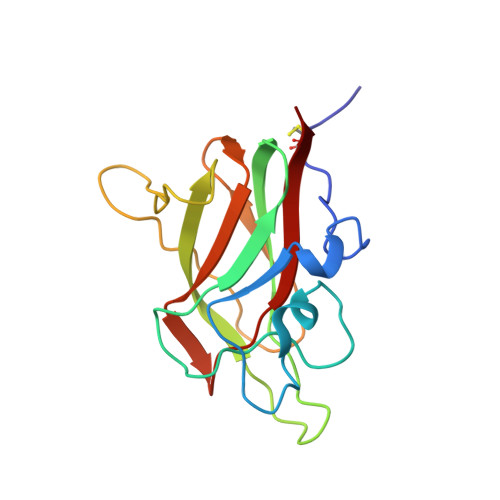
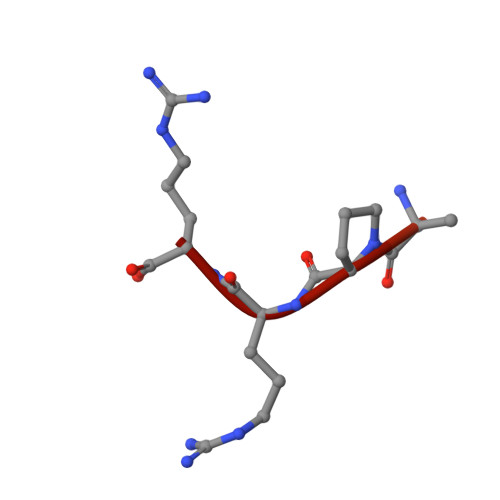
Structural studies of neuropilin-2 reveal a zinc ion binding site remote from the vascular endothelial growth factor binding pocket.
Tsai, Y.C., Fotinou, C., Rana, R., Yelland, T., Frankel, P., Zachary, I., Djordjevic, S.(2016) FEBS J 283: 1921-1934
- PubMed: 26991001
- DOI: https://doi.org/10.1111/febs.13711
- Primary Citation of Related Structures:
5DN2 - PubMed Abstract:
Neuropilin-2 is a transmembrane receptor involved in lymphangiogenesis and neuronal development. In adults, neuropilin-2 and its homologous protein neuropilin-1 have been implicated in cancers and infection. Molecular determinants of the ligand selectivity of neuropilins are poorly understood. We have identified and structurally characterized a zinc ion binding site on human neuropilin-2. The neuropilin-2-specific zinc ion binding site is located near the interface between domains b1 and b2 in the ectopic region of the protein, remote from the neuropilin binding site for its physiological ligand, i.e. vascular endothelial growth factor. We also present an X-ray crystal structure of the neuropilin-2 b1 domain in a complex with the C-terminal sub-domain of VEGF-A. Zn(2+) binding to neuropilin-2 destabilizes the protein structure but this effect was counteracted by heparin, suggesting that modifications by glycans and zinc in the extracellular matrix may affect functional neuropilin-2 ligand binding and signalling activity.
- Institute of Structural and Molecular Biology, University College London, London, UK.
Organizational Affiliation: