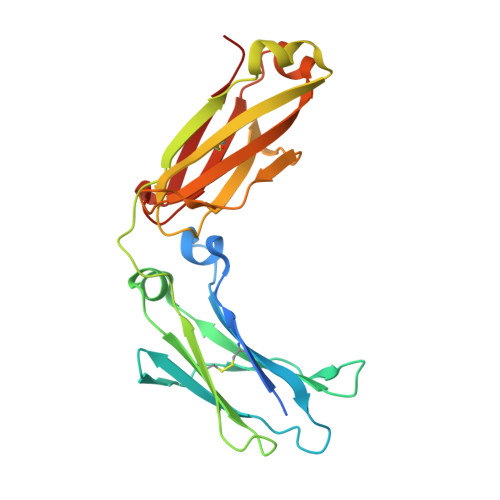
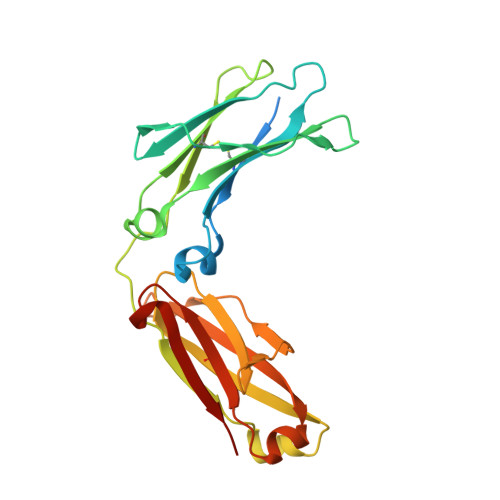
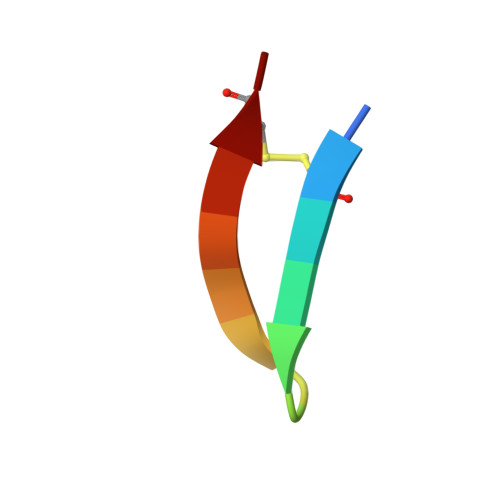
Computationally Designed Bispecific Antibodies using Negative State Repertoires.
Leaver-Fay, A., Froning, K.J., Atwell, S., Aldaz, H., Pustilnik, A., Lu, F., Huang, F., Yuan, R., Hassanali, S., Chamberlain, A.K., Fitchett, J.R., Demarest, S.J., Kuhlman, B.(2016) Structure 24: 641-651
- PubMed: 26996964
- DOI: https://doi.org/10.1016/j.str.2016.02.013
- Primary Citation of Related Structures:
5DI8, 5DJ0, 5DJ2, 5DJ6, 5DJ8, 5DJA, 5DJC, 5DJD, 5DJX, 5DJY, 5DJZ, 5DK0, 5DK2, 5DVK, 5DVL, 5DVM, 5DVN, 5DVO - PubMed Abstract:
A challenge in the structure-based design of specificity is modeling the negative states, i.e., the complexes that you do not want to form. This is a difficult problem because mutations predicted to destabilize the negative state might be accommodated by small conformational rearrangements. To overcome this challenge, we employ an iterative strategy that cycles between sequence design and protein docking in order to build up an ensemble of alternative negative state conformations for use in specificity prediction. We have applied our technique to the design of heterodimeric CH3 interfaces in the Fc region of antibodies. Combining computationally and rationally designed mutations produced unique designs with heterodimer purities greater than 90%. Asymmetric Fc crystallization was able to resolve the interface mutations; the heterodimer structures confirmed that the interfaces formed as designed. With these CH3 mutations, and those made at the heavy-/light-chain interface, we demonstrate one-step synthesis of four fully IgG-bispecific antibodies.
- Department of Biochemistry, University of North Carolina at Chapel Hill, 120 Mason Farm Road, Campus Box 7260, Chapel Hill, NC 27599, USA.
Organizational Affiliation: