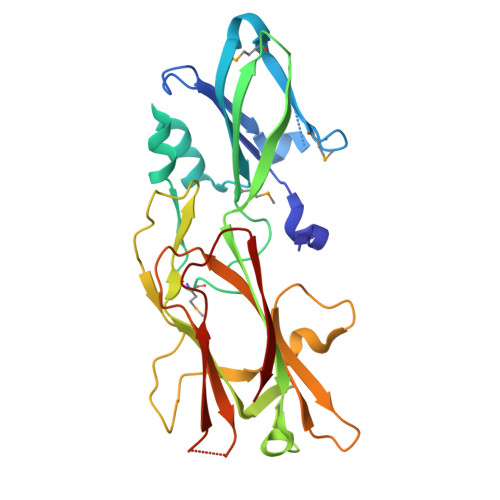
Structure of the fimbrial protein Mfa4 from Porphyromonas gingivalis in its precursor form: implications for a donor-strand complementation mechanism.
Kloppsteck, P., Hall, M., Hasegawa, Y., Persson, K.(2016) Sci Rep 6: 22945-22945
- PubMed: 26972441
- DOI: https://doi.org/10.1038/srep22945
- Primary Citation of Related Structures:
5DHM - PubMed Abstract:
Gingivitis and periodontitis are chronic inflammatory diseases that can lead to tooth loss. One of the causes of these diseases is the Gram-negative Porphyromonas gingivalis. This periodontal pathogen is dependent on two fimbriae, FimA and Mfa1, for binding to dental biofilm, salivary proteins, and host cells. These fimbriae are composed of five proteins each, but the fimbriae assembly mechanism and ligands are unknown. Here we reveal the crystal structure of the precursor form of Mfa4, one of the accessory proteins of the Mfa1 fimbria. Mfa4 consists of two β-sandwich domains and the first part of the structure forms two well-defined β-strands that run over both domains. This N-terminal region is cleaved by gingipains, a family of proteolytic enzymes that encompass arginine- and lysine-specific proteases. Cleavage of the N-terminal region generates the mature form of the protein. Our structural data allow us to propose that the new N-terminus of the mature protein may function as a donor strand in the polymerization of P. gingivalis fimbriae.
- Department of Chemistry, Umeå University, Umeå, SE-901 87, Sweden.
Organizational Affiliation: