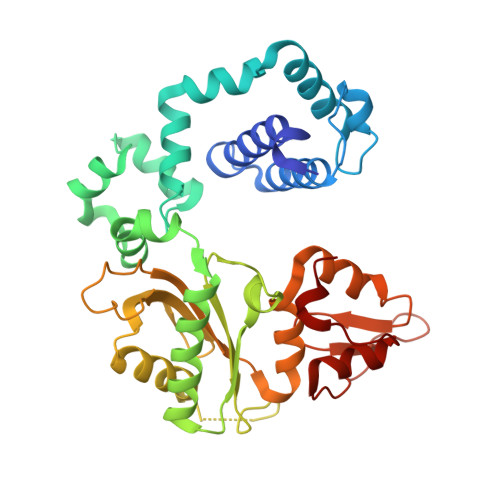
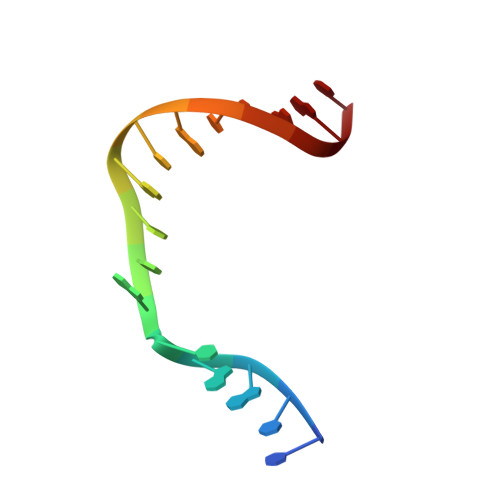
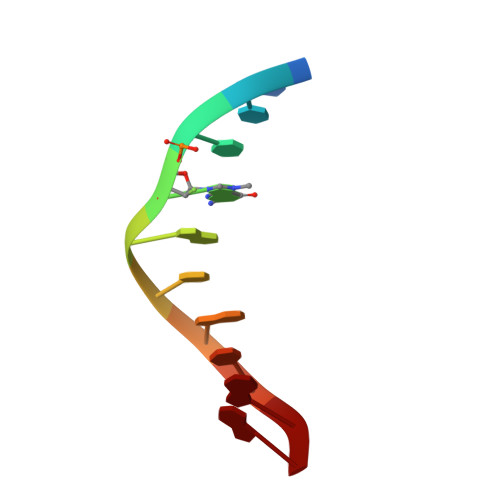
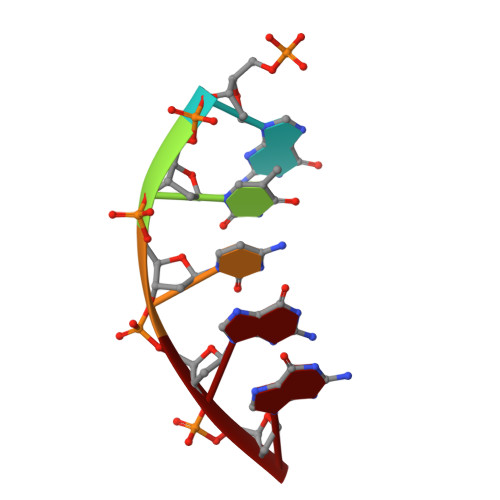
N7 Methylation Alters Hydrogen-Bonding Patterns of Guanine in Duplex DNA.
Kou, Y., Koag, M.C., Lee, S.(2015) J Am Chem Soc 137: 14067-14070
- PubMed: 26517568
- DOI: https://doi.org/10.1021/jacs.5b10172
- Primary Citation of Related Structures:
5DB6, 5DB7, 5DB8, 5DB9, 5DBA, 5DBB, 5DBC - PubMed Abstract:
N7-Alkyl-2'-deoxyguanosines are major adducts in DNA that are generated by various alkylating mutagens and drugs. However, the effect of the N7 alkylation on the hydrogen-bonding patterns of the guanine remains poorly understood. We prepared N7-methyl-2'-deoxyguanosine (N7mdG)-containing DNA using a transition-state destabilization strategy, developed a novel polβ-host-guest complex system, and determined eight crystal structures of N7mdG or dG paired with dC, dT, dG, and dA. The structures of N7mdG:dC and N7mdG:dG are very similar to those of dG:dC and dG:dG, respectively, indicating the involvement of the keto tautomeric form of N7mdG in the base pairings with dC and dG. On the other hand, the structure of N7mdG:dT shows that the mispair forms three hydrogen bonds and adopts a Watson-Crick-like geometry rather than a wobble geometry, suggesting that the enol tautomeric form of N7mdG involves in its base pairing with dT. In addition, N7mdG:dA adopts a novel shifted anti:syn base pair presumably via the enol tautomeric form of N7mdG. The polβ-host-guest complex structures reveal that guanine-N7 methylation changes the hydrogen-bonding patterns of the guanine when paired with dT or dA and suggest that N7 alkylation may alter the base pairing patterns of guanine by promoting the formation of the rare enol tautomeric form of guanine.
- Division of Medicinal Chemistry, College of Pharmacy, The University of Texas at Austin , Austin, Texas 78712, United States.
Organizational Affiliation: