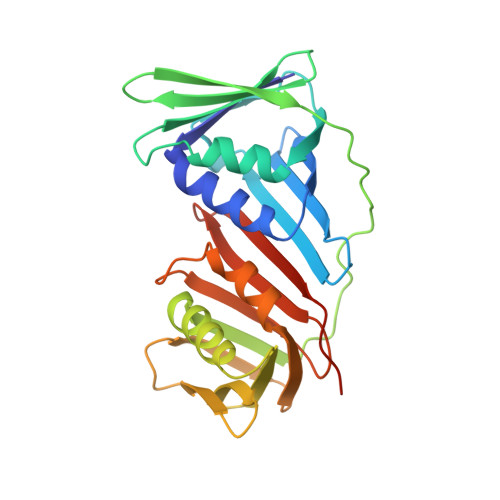
A small protein inhibits proliferating cell nuclear antigen by breaking the DNA clamp.
Altieri, A.S., Ladner, J.E., Li, Z., Robinson, H., Sallman, Z.F., Marino, J.P., Kelman, Z.(2016) Nucleic Acids Res 44: 6232-6241
- PubMed: 27141962
- DOI: https://doi.org/10.1093/nar/gkw351
- Primary Citation of Related Structures:
5DA7, 5DAI - PubMed Abstract:
Proliferating cell nuclear antigen (PCNA) forms a trimeric ring that encircles duplex DNA and acts as an anchor for a number of proteins involved in DNA metabolic processes. PCNA has two structurally similar domains (I and II) linked by a long loop (inter-domain connector loop, IDCL) on the outside of each monomer of the trimeric structure that makes up the DNA clamp. All proteins that bind to PCNA do so via a PCNA-interacting peptide (PIP) motif that binds near the IDCL. A small protein, called TIP, binds to PCNA and inhibits PCNA-dependent activities although it does not contain a canonical PIP motif. The X-ray crystal structure of TIP bound to PCNA reveals that TIP binds to the canonical PIP interaction site, but also extends beyond it through a helix that relocates the IDCL. TIP alters the relationship between domains I and II within the PCNA monomer such that the trimeric ring structure is broken, while the individual domains largely retain their native structure. Small angle X-ray scattering (SAXS) confirms the disruption of the PCNA trimer upon addition of the TIP protein in solution and together with the X-ray crystal data, provides a structural basis for the mechanism of PCNA inhibition by TIP.
- Institute for Bioscience and Biotechnology Research, University of Maryland and the National Institute of Standards and Technology, 9600 Gudelsky Drive, Rockville, MD 20850, USA.
Organizational Affiliation: